NCBI ClinVar
The NCBI ClinVar database collects reports from scientists on human genetic variants and their relationships to disease. Its purpose is to bridge genetic variation data and observations or assertions about phenotypes that are related to those variations.
You can search ClinVar with gene symbols, diseases, HGVS expressions, location on a chromosome, and more.
This tutorial demonstrates how to find specific disease-causing genetic variants.
Click Next to begin.
ClinVar Search
1 of 2
Over the years, scientists have named gene variants in many ways. NCBI has attempted to account for these various names by including them in the NCBI database records. Search ClinVar by the information you have and use the database to uniquely identify the variant of interest.
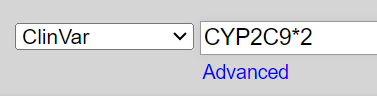
Let’s imagine that we were asked to find information about a variant described as "CYP2C9*2."
The first part of this string resembles a gene symbol, so we’ll take a guess that CYP2C9*2 is a variant of the CYP2C9 gene. We'll search ClinVar for CYP2C9*2 and see if we can find a record that matches.
Search: CYP2C9*2
Note: The asterisk (*) is not a wildcard in ClinVar.
ClinVar Search
2 of 2
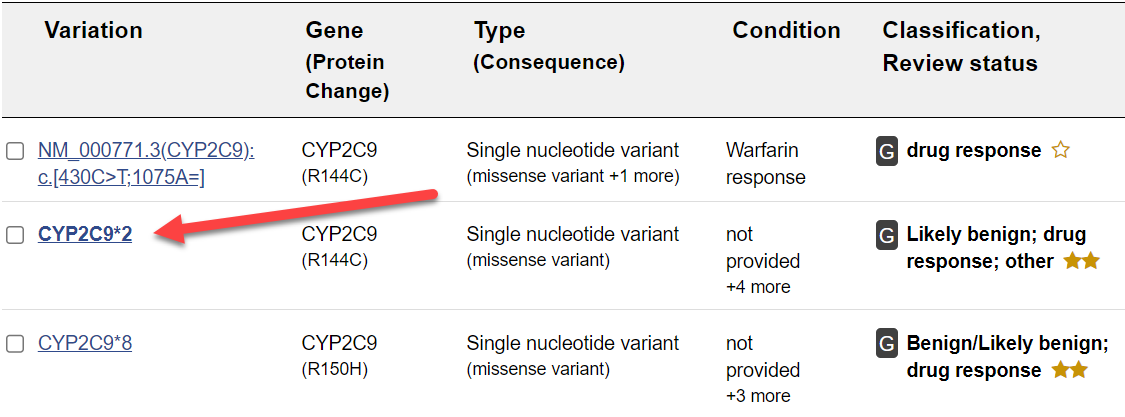
You should see 10 or so records in your results for the search of CYP2C9*2 in ClinVar (your results may vary slightly).
This results table provides basic information about the variation, the related gene(s), the type or molecular consequence, associated conditions, clinical significance, and the review status of the assertion that the genetic variation and the condition or phenotypic observation are related.
Let's look at the best matching record in more detail.
Click on the link labeled CYP2C9*2.
ClinVar Record
1 of 4
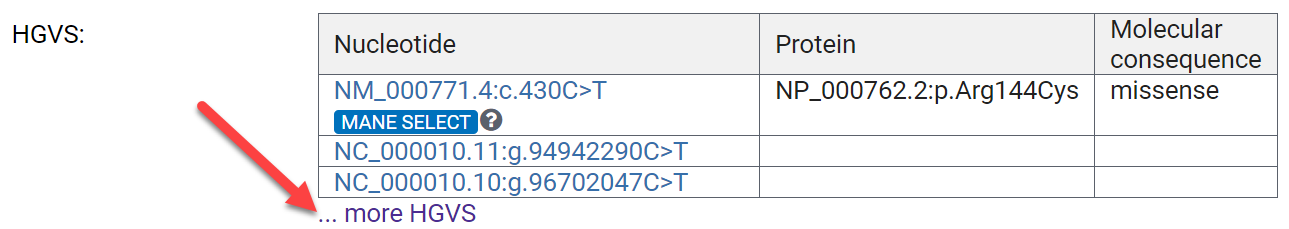
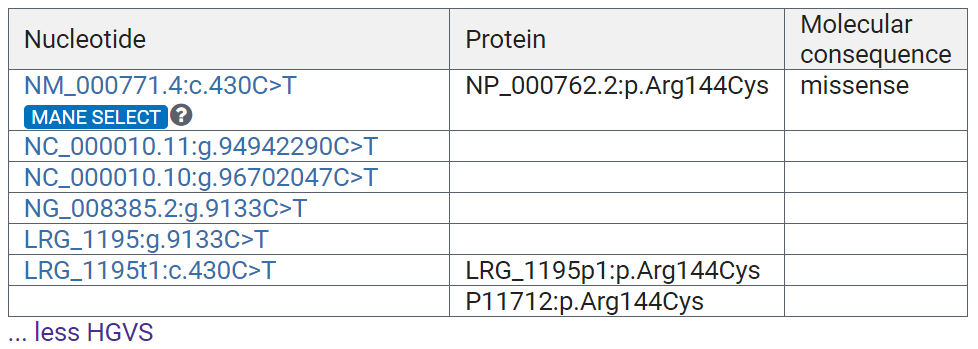
Let's first focus on the Variant Details, on the table labeled HGVS. These are Human Genome Variation Society (HGVS) expressions for this variation.
Expand the HGVS section by clicking ...more HGVS.
ClinVar Record
2 of 4
Most ClinVar record titles are in HGVS nomenclature. This record happens to have a different title.
Let's break down how to read the HGVS expression.
Under Nucleotide starting NM:
- NM_000771.4 refers to the reference sequence transcript.
- c.430 is the nucleotide position in the coding region of the reference transcript.
- C>T means that the C at position 430 of the coding region of NM_000771.4 is changed to a T.
- MANE SELECT are high quality transcripts, where the reference sequence from NCBI and the one from the European Molecular Biology Laboratories-European Bioinformatics Institute (EMBL-EBI) are identical.
Under the Protein column:
- NP_000762.2 refers to the protein reference record.
- p.Arg144Cys refers to a resultant amino acid change in the protein.
HGVS expressions are standardized nomenclature for describing variations, so are incredibly useful in communicating about genetic variation.
ClinVar Record
3 of 4
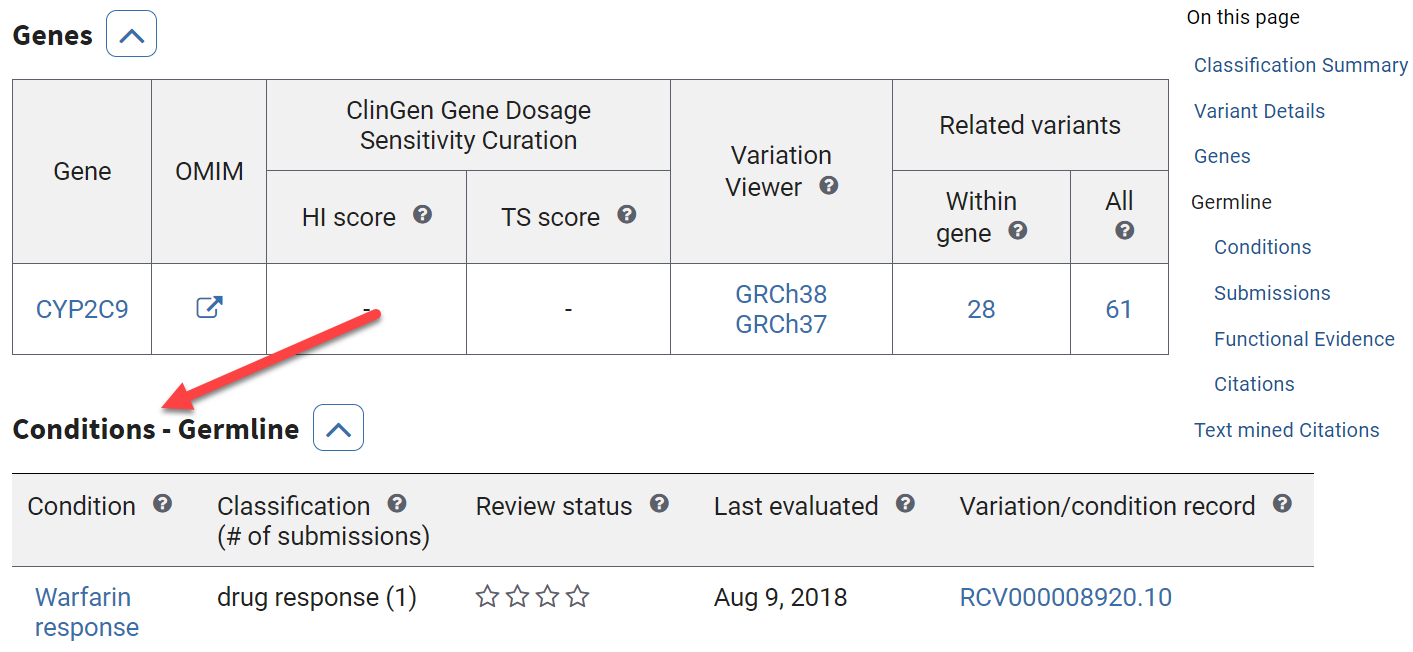
Scroll down to the Conditions section.
There are only Germline related conditions. If there were somatic variations associated with conditions, you would see another table here.
What clinical significance is associated with this variation in the CYP2C9 gene?
Correct!
Submitters have suggested that this variant is associated with responses to the drugs piroxicam, flurbiprofen, lesinurad, warfarin, and phenytoin.
Incorrect!
Please try again.
Incorrect!
Please try again.
Incorrect!
Please try again.
ClinVar Record
4 of 4
Scrolling back up to the top of the record, note that ClinVar reports the level of review supporting the assertion of clinical significance for the variation under Classification.
The stars reflect the level of agreement on the clinical assertion.
You can view the specific definition of each review status in the ClinVar documentation.
What do the number of gold stars (two) on this particular record mean?
Incorrect!
Please try again.
Incorrect!
Please try again.
Correct!
That is correct.
Incorrect!
Please try again.
Conclusion
Congratulations! You can now find specific disease-causing genetic variants using ClinVar.
You can now close the NLM Navigator windows.