NCBI Nucleotide
The NCBI Nucleotide Database is a database of nucleic acid sequences. These sequences come from laboratories around the world that submit their data to one of a set of repositories, including GenBank, which is maintained by NCBI.
Other records are "Reference Sequences," which are representative (model) examples of sequences, curated by NCBI.
Where possible, the sequences are annotated so that you can find the strings of sequences that may be functional.
Let's look at examples so that you can become familiar with the parts of a Nucleotide record.
To begin, please click the Next button below.
Example 1: Giant Viruses
1 of 6
You might have read in the news or heard on the radio or TV about the new giant viruses that have been discovered.
Skim this Science article abstract and find the name for some of the viruses.
Example 1: Giant Viruses
2 of 6
Now try searching the NCBI Nucleotide database for klosneuvirus.
Results appear in the middle of the page. Search filters and other discovery tools appear on either side of results.
Each result in the database represents a record for one sequence or string of nucleic acids represented as base pairs ("bp" as shown in the results). The strings may overlap.
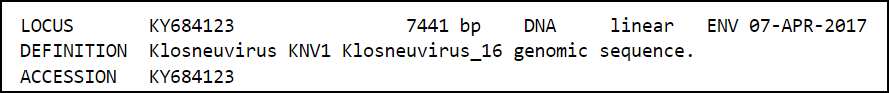
Click on the record for accession number KY684123.1 and view the full record.
[Note: Sidebar images like the above can be expanded by clicking on them.]
Example 1: Giant Viruses
3 of 6
Look at the record for Klosneuvirus 16 and answer the following questions. For help, view a Sample GenBank record.
What is the sequence length or number of base pairs?
Correct!
There are 7441 base pairs (or "bp") in this sequence. You can see that in the line labeled "LOCUS" and you may also have seen that on the results screen in the summary display.
Incorrect!
Oops! Are you looking at the record for Klosneuvirus KNV1 Klosneuvirus_16 genomic sequence?
Incorrect!
Remember that base pairs are also labeled "bp."
HINT: This information is stored in the Locus field.
Example 1: Giant Viruses
4 of 6
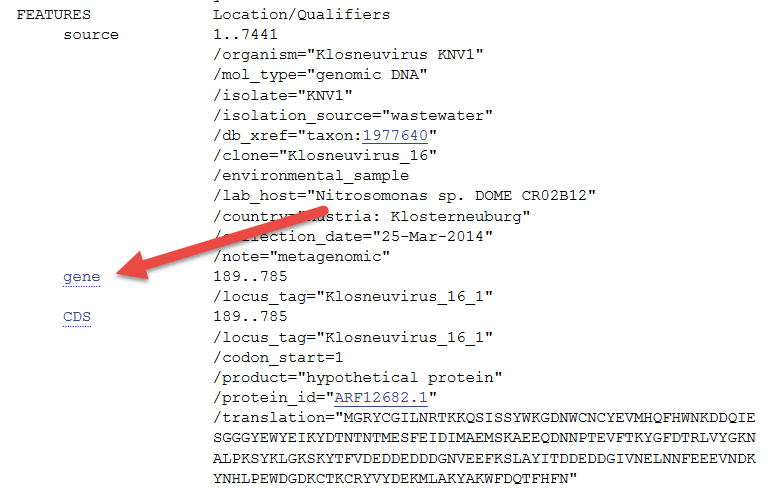
Scroll down the feature table and click on the first link for gene.
This link "jumps" you to the raw sequence data at the bottom of the record, highlighting the part of the sequence identified as a gene.
New navigation features appear at the bottom of your screen, allowing you to jump from feature to feature.
How many genes are annotated (defined) on this accession number?
Incorrect!
No, see the number of sections marked "Gene."
Incorrect!
No, see the number of sections marked "Gene."
Correct!
That is correct.
HINT: This information is included on the bottom of the page.
Example 1: Giant Viruses
5 of 6
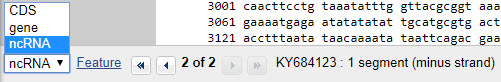
Now scroll back up to the feature table and notice how the different features ("gene," "CDS," "ncRNA") are annotated. See what features have products.
What are the first ten nucleotides in the HNH endonuclease gene?
Correct!
That is correct.
Incorrect!
No, this is the translation to amino acids. The nucleotides are highlighted when you click on the CDS link.
Incorrect!
No, somehow you've come across the wrong sequence. Find the nucleotides corresponding to the gene by clicking on the CDS link in the feature table to highlight the correct part of the code.
HINT: Look through the feature table to find a part of the sequence with a product labeled, "HNH endonuclease." Follow the link in the feature table to the sequence at the bottom of the record to see the first ten nucleotides.
Example 1: Giant Viruses
6 of 6
Are any RNA molecules annotated (defined) on this record (accession number KY684123)?
Correct!
The RNA segments are labeled "ncRNA" in the feature table.
Incorrect!
Look again. The RNA segments are labeled "ncRNA" in the feature table.
HINT: Either look through the feature table for features marked "RNA" or check the drop down menu at the bottom of the screen.
"ncRNA" stands for "non-coding RNA." It is a sequence that may regulate gene expression, but doesn't code for a protein.
Example 2: Mitochondrial Transporter SLC25A3
1 of 5
Start a new search at the top of the screen.
Search for SLC25A3.
The protein encoded by this gene catalyzes the transport of phosphate into the mitochondrial matrix, either by proton cotransport or in exchange for hydroxyl ions.
Use your search results from Nucleotide to answer the following questions about SLC25A3:
Approximately how many sequences exist in the NCBI Nucleotide database for the mitochondrial transporter SLC25A3?
Incorrect!
No, we're looking for the total number of records that result from a search of SLC25A3 in the Nucleotide database.
Correct!
We're looking for the total number of records that result from a search of SLC25A3 in the Nucleotide database.
Incorrect!
No, we're looking for the total number of records that result from a search of SLC25A3 in the Nucleotide database.
Example 2: Mitochondrial Transporter SLC25A3
2 of 5
Approximately how many sequence records in your results are from the INSDC (GenBank)?
Incorrect!
The filter labeled "INSDC (GenBank)" shows the number of your results that are sequences from INSDC sources.
Incorrect!
The filter labeled "INSDC (GenBank)" shows the number of your results that are sequences from INSDC sources.
Correct!
That is correct.
HINT: Find your filter options on the left of your screen.
The International Nucleotide Sequence Database Collaboration (INSDC) is a joint effort to collect and disseminate annotated DNA and RNA sequences. The collaboration includes NIG's DNA Data Bank of Japan, NCBI's GenBank and the EMBL-EBI's European Nucleotide Archive.
Example 2: Mitochondrial Transporter SLC25A3
3 of 5
Approximately how many of your search results are RefSeq records?
Incorrect!
Look at the filter under Source Databases labeled "RefSeq."
Incorrect!
Look at the filter under Source Databases labeled "RefSeq."
Correct!
That is correct.
RefSeq is short for the NCBI Reference Sequence collection which is a comprehensive, integrated, non-redundant, well-annotated set of sequences, including genomic DNA, transcripts, and proteins. RefSeq sequences form a foundation for medical, functional, and diversity studies.
Example 2: Mitochondrial Transporter SLC25A3
4 of 5
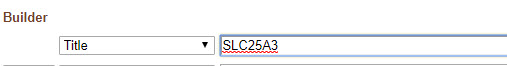
Now start a new search using the Advanced page (making sure to clear any applied filters first).
Search for SLC25A3 in the Title field.
You can use the Add to history link to stay on the Advanced page and compare your results.
How much of a difference is there between the results for this search and your last one?
Incorrect!
Please try again.
Correct!
That is correct.
Incorrect!
Please try again.
HINT: Compare the following searches:
- SLC25A3
- SLC25A3[title]
Example 2: Mitochondrial Transporter SLC25A3
5 of 5
Return to your search results for your SLC25A3 search (WITHOUT the [title] field restriction).
Are there any SLC25A3 sequences in plants?
Correct!
There are at least 15 sequences in plants.
Incorrect!
Try filtering by Species - Plant.
HINT: Check the Species filters in the left column of your results.
Example 3: Human Tyrosine Hydroxylase
1 of 3
Start a new search at the top of the screen.
Find human tyrosine hydroxylase gene sequence records.
This gene can cause Segawa syndrome and is important in the synthesis of catecholamines (which are important pharmacologic agents).
What is the gene symbol or abbreviation for tyrosine hydroxylase?
Answer: The official gene symbol for tyrosine hydroylase is TH. This is included in most titles of Nucleotide records.
A place to check official gene names is the HGNC, or HUGO Gene Nomenclature Committee (https://www.genenames.org/).
Example 3: Human Tyrosine Hydroxylase
2 of 3
Look at the following accession records, all of which are in your search results for the tyrosine hydroxylase (TH) gene:
- L15440.1
- X05290.1
- M17589.1
How are these records different from each other?
Look at their sizes, their sources, any descriptive notes, and their feature tables.
HINT: These three records are in your search results, but for easier comparison search Nucleotide for: L15440.1 OR X05290.1 OR M17589.1
Example 3: Human Tyrosine Hydroxylase
3 of 3
Each of the records you looked at included a sequence that codes for the TH gene.
L15440.1: The record title says it all. This records a human DNA sequence that includes several genes, not just TH. And it only has the 3' end of the TH gene. See the top of the feature table, and you can see that base pairs 1 through 1482 are part of the TH gene.
X05290: This records an mRNA sequence that encodes a single protein product, identified as "tyrosine hydroxylase (HTH-1)." Note that it includes only about 1,800 base pairs. In the CDS portion of the feature table, note that the protein product is called HTH-1.
M17589.1: The feature table on this record includes only one gene and one CDS region. Note that the literature reference says that "alternative RNA splicing produces four kinds of mRNA from a single gene" and the annotation in the feature table says that this one is type 4.
Conclusion
1 of 2
The Nucleotide database is a database of nucleic acid sequences. These sequences come from laboratories around the world that submit their data to one of a set of repositories, including GenBank, which is maintained by NCBI. Other records are "Reference Sequences," which are representative (model) examples of sequences, curated by NCBI.
As you have seen in the tutorial, you can look up nucleotide sequences a variety of ways.
Conclusion
2 of 2
Congratulations! You've completed this exercise in exploring NCBI Nucleotide.
You can now close the NLM Navigator windows.