NCBI Protein: Simple Search and Record Structure
The NCBI Protein database is a database of protein (amino acid) sequences.
Some of the sequences in the Protein database come from laboratories that have done protein sequencing. This is called "primary" data. Some of the sequences are derived from genetic sequences, such as the NCBI RefSeq (or Reference Sequence) records. This is called "derived" data.
Let's look at an example to explore the Protein database. Click Next to begin.
Finding a Protein Sequence Record
1 of 4
We will start with a simple search: How many sequences exist in the Protein database for the mitochondrial transporter SLC25A3?
Check that Protein is selected from the database selection menu, enter SLC25A3 and click Search.
Finding a Protein Sequence Record
2 of 4
You should have retrieved more than 1400 records.
Explore the filters on the left to learn what kinds of sequences the Protein database contains.
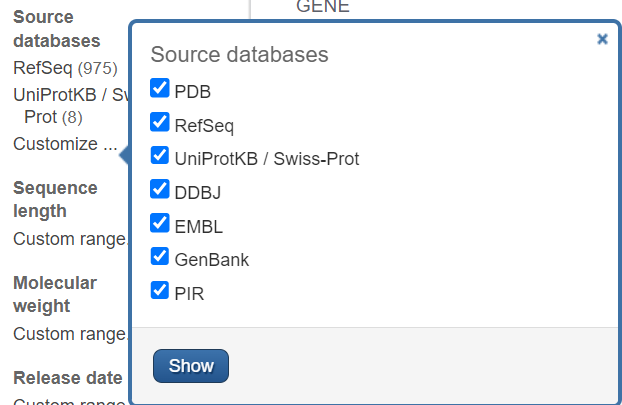
Under Source databases, use the Customize link to view all of the sources of protein records in this database. You can select those you wish to view, then click Show to have them appear on your results page.
PDB records and UniProt records may be primary sequence data.
Most of the records for SLC25A3 are RefSeq records, which are derived from nucleotide sequence data.
Finding a Protein Sequence Record
3 of 4
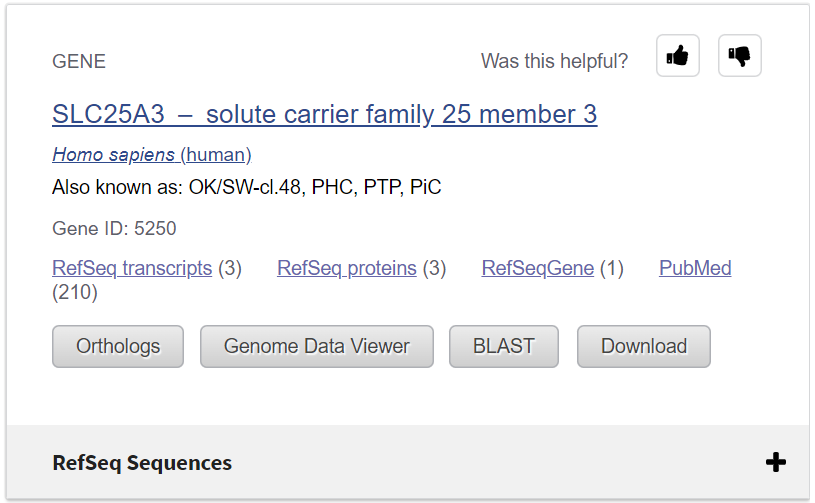
At the top of your results is a quick link to the relevant record in the Gene database. This appears when your search matches a gene name or symbol.
Under the link to the Gene record, there are buttons to other resources and an expandable section called "RefSeq Sequences."
Let's explore the Protein reference sequences for this gene.
Click to expand RefSeq Sequences +.
Finding a Protein Sequence Record
4 of 4
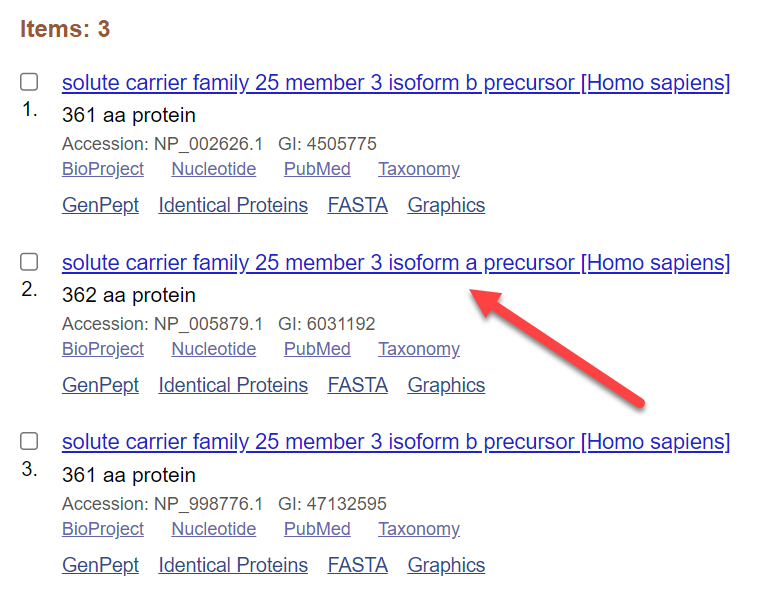
You should now see three rows that include links to three reference sequences in the Protein database. Different splice variants will be represented by different protein records.
For SLC25A3, there is one sequence for isoform a (with 362 amino acids) and two sequences that result in isoform b (with 361 amino acids).
Click to view the record for NP_005879.1 (Isoform a).
Understanding a Protein Sequence Record
1 of 2
Take a moment to familiarize yourself with this protein reference sequence record. This is a record that is curated by NCBI.
The following are some notes and questions about the record:
Below the title, the label of "NCBI Reference Sequence" and record prefix "NP_" identify this as an NCBI protein reference sequence record.
The DBSOURCE field tells you what genomic reference sequence this protein sequence is derived from (NM_005888.4).
The ORGANISM field lists the complete taxonomic hierarchy of that organism.
What organism is this protein from?
Incorrect!
Please try again.
Correct!
That is correct.
Incorrect!
Please try again.
Understanding a Protein Sequence Record
2 of 2
In the list of literature references, note the specific sequence of residues discussed, the full citation data, the link to PubMed, and "REMARK," which notes the findings of the research cited.
The COMMENT field summarizes what is known about the function of this protein and relevant information about transcript variants.
What is the difference between variant 1 and variant 2 of this protein?
Variant 1 has an alternate exon in the 5' coding region. The resultant protein (isoform a) has an internal segment that is different from isoform b.
The FEATURES table points out (or "annotates") specific segments, sites or regions of the sequence and their known products or functions. Clicking on a link in this table will highlight the amino acids (or "residues") involved in the sequence at the bottom of the record.
Which of the following site types have been annotated in this protein?
Incorrect!
Please try again.
Incorrect!
Please try again.
Incorrect!
Please try again.
Incorrect!
Please try again.
Correct!
That is correct.
The ORIGIN section at the bottom of the record is the actual sequence of amino acids or "residues."
Explore this record to your satisfaction.
Conclusion
Congratulations! You have completed this tutorial on the NCBI Protein database search and record structure.
You can now close the NLM Navigator windows.