Supplementary Information
Supplementary Information
In this section you will find additional information on some popular topics that we did not have time to cover in depth during the workshop.
Instructions for AWS Glue – AWS Athena is a powerful tool to query information in a database. However, importing a database can be challenging. AWS Glue is another cloud tool that can be used to quickly import data tables into Athena for use. The instructions in this section will walk you through the setup of the SRA data tables for use with Athena.
SQL Challenges – SQL is the required language for generating queries in the AWS Athena service. Today’s workshop gave you an introduction to its capabilities, but if you are interested in a deeper understanding, you can visit this section to learn additional skills and try your hand at some challenges using the SRA data tables.
Instructions for AWS Glue
In order search through a table (like the SRA metadata table) you need to load it into Athena. Because this is your first-time accessing Athena, it shouldn’t be a surprise that you don’t currently have any data tables loaded! Therefore, our first step will be to add the SRA metadata table to Athena. There are three ways you can add a table to Athena:
- Create the table using SQL commands (we are not SQL experts here…yet, so we won’t do it this way)
- Add the table manually from an S3 bucket (although the SRA metadata is stored in its own S3 bucket, we don’t know the format of the table, so we can’t do it this way)
- Use another AWS service called AWS Glue to automatically parse a table and load it into Athena for us using a “Crawler” (this is what we will do
Note: Although AWS Glue is the most convenient method, it is also the only one to cost money. To parse the SRA metadata table it will be… ~$0.01.
This section will walk through the steps taken during the AWS Glue demo to prepare the SRA metadata table for Athena queries
1) To start working with AWS Glue, navigate to the Tables section of the Athena page on the left-hand side, then click Create Table and select from AWS Glue Crawler as seen below. If you see a pop-up about the crawler, just click Continue.
2) The settings for this crawler should e set as described below:
- Crawler name: This name needs to be informative, but not universally unique (like an S3 bucket). Choose something that helps you remember what the crawler is trying to access.
Next page
- Crawler Source Type: Keep the default setting (data stores)
- Repeat crawls of S3 data stores: Keep the default setting (Crawl all folders)
Next page
- Choose a data store: Set it to S3
- Connection: Leave empty
- Crawl data in: Select Specified path in another account
- Include path: This is the path to the table itself. The SRA metadata table used here is located at s3://sra-pub-metadata-us-east-1/sra/metadata/
Next page
- Add another data store: Select No. However, crawlers can parse multiple tables at a time, so you can add this if you want.
Next page
- Choose an IAM role: Work with your admin to determine the correct choice for you. If you are doing this on your own, I recommend creating your own IAM role and reusing it if you need additional crawlers.
Next page
- Frequency: Select Run on Demand. Crawlers can be run on specified intervals, but this costs extra money, and for most purposes is unecessary.
Next page
- Database: Select Add database then make a name for it. The tables parsed by this crawler will be stored in this database. Then click Create.
- Prefix added to tables (optional) - Leave empty.
Next Page
Click Finish
3) You should now be on the Crawler page for AWS Glue. Here you can manage and run crawlers. Click the checkbox next to the new crawler and select Run Crawler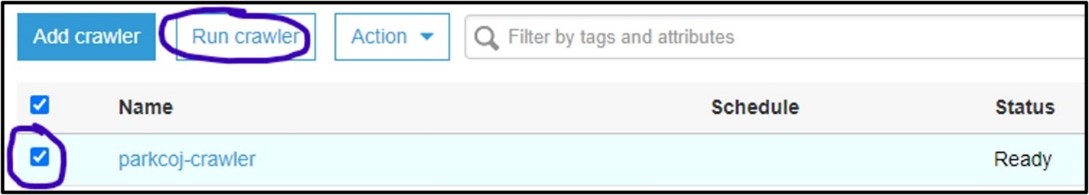
4) If it worked, you should see the Status column say Ready again, and the Tables added column should have changed to 1:
5) The table should now appear in Athena and you can follow the same steps described in section Exploring Athena Tables above.
SQL Challenges
Now that you have a handle on how SQL commands work, let’s try some examples! Remember, you can use the “Tables” section on the left-hand side of the page to find out which columns you can filter by in your table.
If you find yourself stumped on the answer to any of these, or just want to check your answer, click the dropdown box underneath the question to reveal the solution and see a screenshot of the results table!
a) You just came across a new paper with lots of great sequence data. You want to add that data to your own research so you jump to the paper’s Data Availability section (because all great computational papers have one!) and see that the data was stored in an SRA study under the accession SRP291000. Write a SQL command in the query terminal to find all data associated with the SRA study accession SRP291000:
Click here to see the solution!
SELECT * FROM "sra"."metadata" WHERE sra_study = 'SRP291000'
b) You are working on a new genome assembly tool for short-read sequences. However, you don’t have any reads of your own to test it! You know that SRA metadata includes the sequencing platform reads were generated on, so you decide you want to check there. Write a SQL command in the query terminal to find all data sequenced with the OXFORD_NANOPORE platform.
Click here to see the solution!
SELECT * FROM "sra"."metadata" WHERE platform = 'OXFORD_NANOPORE'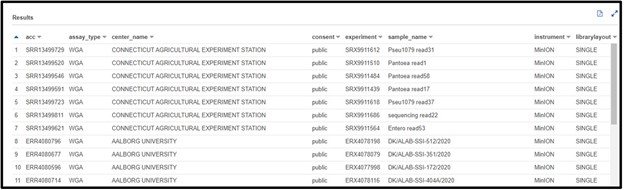
Now let’s get a little bit more complicated with our queries by combining multiple filtering conditions together. For example, see the command below: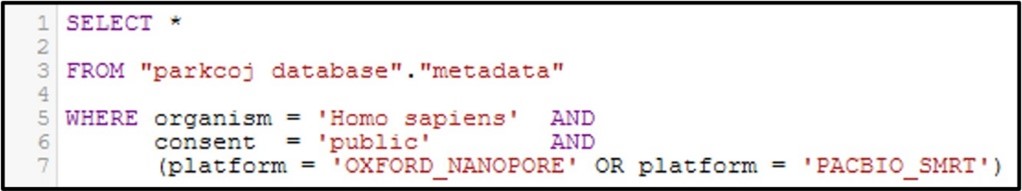
In this command we use the AND statement to add multiple requirements for our data. Specifically, we added a second criteria where the consent = public (aka: The data is not under restricted access). Additionally, we add a more complex requirement by using an OR statement for the platform column to ask for data that was generated by the OXFORD_NANOPORE OR PACBIO_SMRT sequencing platforms. Overall, by running this command we will only get the data that fits all 3 conditions.
Note: Make sure you include parenthesis around an OR statement as seen above, otherwise the query may not work as intended.
Here’s a few brain teasers to flex your new SQL skills! Remember, if you are stuck you can click the dropdown box underneath the question to reveal the solution and see a screenshot of the results table!
a) After testing your genome assembly tool from earlier, you realize that not all Illumina datasets are created equally! It turns out you only need WGS (Whole Genome Sequencing) genomic data to properly validate your software. Also, you noticed that there was some metagenomic and transcriptomic data mixed in with your test cases. So, this time you are just going to look for “genomic” datasets. Write a SQL command in the query terminal to find all WGS assay_typedata sequenced on the ILLUMINA platform and a GENOMIC library_source.
Click here to see the solution!
SELECT * FROM "sra"."metadata" WHERE platform = 'ILLUMINA' AND assay_type = 'WGS' AND librarysource = 'GENOMIC'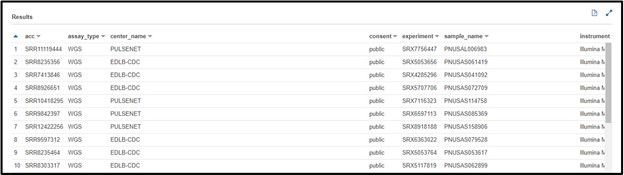
b) You are designing a population-level epidemiological survey of some bacterial pathogens from samples collected across Europe. You decide you want to get some preliminary data on Escherichia coli(or maybe Staphylococcus aureus…) from the SRA, but you aren’t picky about what kind of sequencing is done just yet. Write a SQL command in the query terminal to find all sequences collected from the continent Europe and are from the organism Escherichia coli or Staphylococcus aureus.
Hint: The column header for the continent is not very intuitive. Try using the “Preview Table” option from the “Tables” tab described earlier to find a column that would fit.
Click here to see the solution!
SELECT * FROM "sra"."metadata" WHERE (organism = 'Escherichia coli' OR organism = 'Staphylococcus aureus') AND geo_loc_name_country_continent_calc = 'Europe'
Page 1 of 1
Last Reviewed: July 14, 2022

