How I develop "human health" examples....
How we develop relevant examples with NCBI resources

-
-
-
- Who is the audience?
- Clinical Students & Clinicians (also biology students & translational science researchers)
- What do they need to learn?
- The clinical and molecular ramification of their patients' genetic variations
- What do they need to learn how to do?
- Learn about an unfamiliar genetic disorder/disease
- Find a relvant genetic test
- Retrieve information about the patient's genetic variant
- Learn about the impacted gene
- Map the variant onto the biomolecules produced from that gene
- Interpret & integrate the information to explain the patient's symptoms
- Be able to explain what is happening in the patient & their family
- What is a relevant and interesting story for the audience?
- Real patient cases!
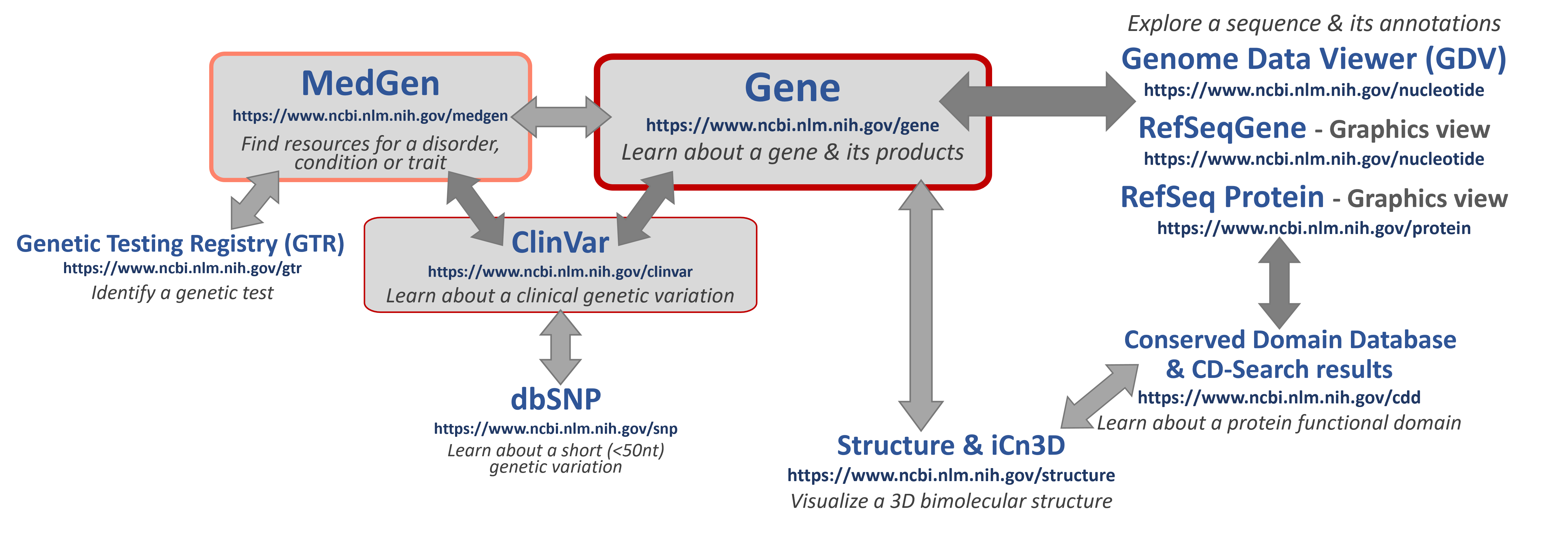
- What kind of NCBI data & resources are relevant?
- Clinical resources
- MedGen
- GTR
- ClinVar
- Biological Sequence resources
- Gene
- Nucleotide
- Protein
- Genome Data Viewer & Sequence Viewer (Graphics)
- Biological structure resources
- Conserved Domains database & CD-Search view
- Structure
- iCn3D
- Clinical resources
- What workflow will help find the data and provide the information to solve the case study?
- Workflow: See below
- How can they assess that they've mastered the content?
- Multiple choice questions in a practice (summative) & scored (formative) exam
- Practice with other patient cases with & without guidance
- Who is the audience?
-
-
Tips & Tricks for developing these exercises

A data searching "trick"
Workflows to solve a genetic disease/disorder case
A jump-through for an example case!
- WHAT IS THE QUESTION?
- Where do I start? (Begin with finding relevant background information) Researching the referral
- Understanding the information. (Understanding the retrieved data) Understanding the genetic test results
- Finding more key information. (Linking to select, related data) Learning about the implicated gene
- Integrating the information. (Interpreting the data in context) Mapping the variant & predicting the biological impact
- ANSWERING THE QUESTION
- There is a practice example with guidance available for them to test their skills.
- There are also sets of patient case forms available for futher testing skills without guidance.
How might you adapt this approach for your specific audience?
Welcome to Your Patient!
⇑Researching the Referral
-
-
- To learn more about a case, please click on the Referral icon above to open the form.
- Read it over and fill in below what you can glean from the description and proposed preliminary diagnosis.
-
| Notes | |
| Phenotype | |
| Preliminary Diagnosis |
Note: MedlinePlus® is a service run by the National Library of Medicine for patients and their families and friends. The mission is to to present high-quality, relevant health and wellness information that is trusted, easy to understand, and free of advertising, in both English and Spanish. (learn more)
What does the patient's genetic test report say?
In this scenario, the genetic test was selected and performed. The results have come back from the laboratory and have been sent to you for consultation.-
-
- To learn more about Bo's genetic test results, please click on the Test Results icon above to open the form.
- Read it over and fill in below what you can glean from the report with regard to any genetic variants found and anything they laboratory might be asserting about how this relates to the preliminary diagnosis.
-
 If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Notes | |
| Genetic Variation(s) | |
| Laboratory Assertion(s) |
⇑
What is currently known about the identified variant?
Genetic testing laboratories attempt to stay up to date with what is known about the genetic variants that they are assessing. However, it is sometimes valuable to quickly consult with national database of clinical variants (NCBI's ClinVar database) to learn what other organizations have asserted/interpreted for that variant, if anything. In addition to information from testing laboratories, ClinVar receives curated interpretations from authoritative sources such as ClinGen, ACMG and disorder-specific specialist panels. If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Notes on F9:p.Asp110Gly | |
Variant Information:
|
|
-
- Clinical Interpretation
- Does this match what was listed on the lab report?
- Where on this record can you find out who submitted assertions and what evidence they pointed to?
- HGVS (Human Genome Variation Syntax) aliases, in particular - look for the NG_ and the first NP_ accession-located variants.
- These will be helpful for mapping the variant on these structures and sequences.
- dbSNP Link (and rsID) - The dbSNP database has been collecting biological genetic variant data for 25 years!
- Human population data has been added to relevant variant records from several major projects. What about your patient's variant?
- Clinical Interpretation
⇑
If a particular gene has been implicated in a genetic test results indicating a pathogenic variant exists in a patient, it is often helpful to understand what that gene is, what its normal function is, where it is found (cellular and tissue expression patterns), and other sources of accessible information, such as links to relevant scientific literature.
NCBI's Gene database aggregates data from many NCBI databases as well as other high-quality resources to provide information and links to help users find and understand what is currently known about a particular organism's gene.
 If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Notes | |
Gene Information on the NCBI Gene record:
|
|
-
-
- To learn more about Bo's impacted gene, search NCBI's Gene database with the gene symbol indicated on the genetic test result and find the record for the human version.
- Look over the Gene record that was implicated in the genetic test result and identify:
-
-
- Summary
- What does this gene normally do? What else has been noted?
- Expression
- In which tissues is this gene normally expressed? (Note there are several tissue sets from different project available to view. Try selecting them in the pull-down menu.)
- Gene Ontology project information
- Where in the cell does this gene product act?
- In what processes/pathways?
- Is there more specific confirmation about what it does?
- Summary
⇑
Map the variant through the bioinformatic flow!
Now that we understand which gene may be affected by the presence of the detected variant, mapping the variant through the central dogma of molecular biology can help indicate at which point it has its strongest impact.
 Click here to review an overview of the central dogma and genetic variation.
Click here to review an overview of the central dogma and genetic variation.
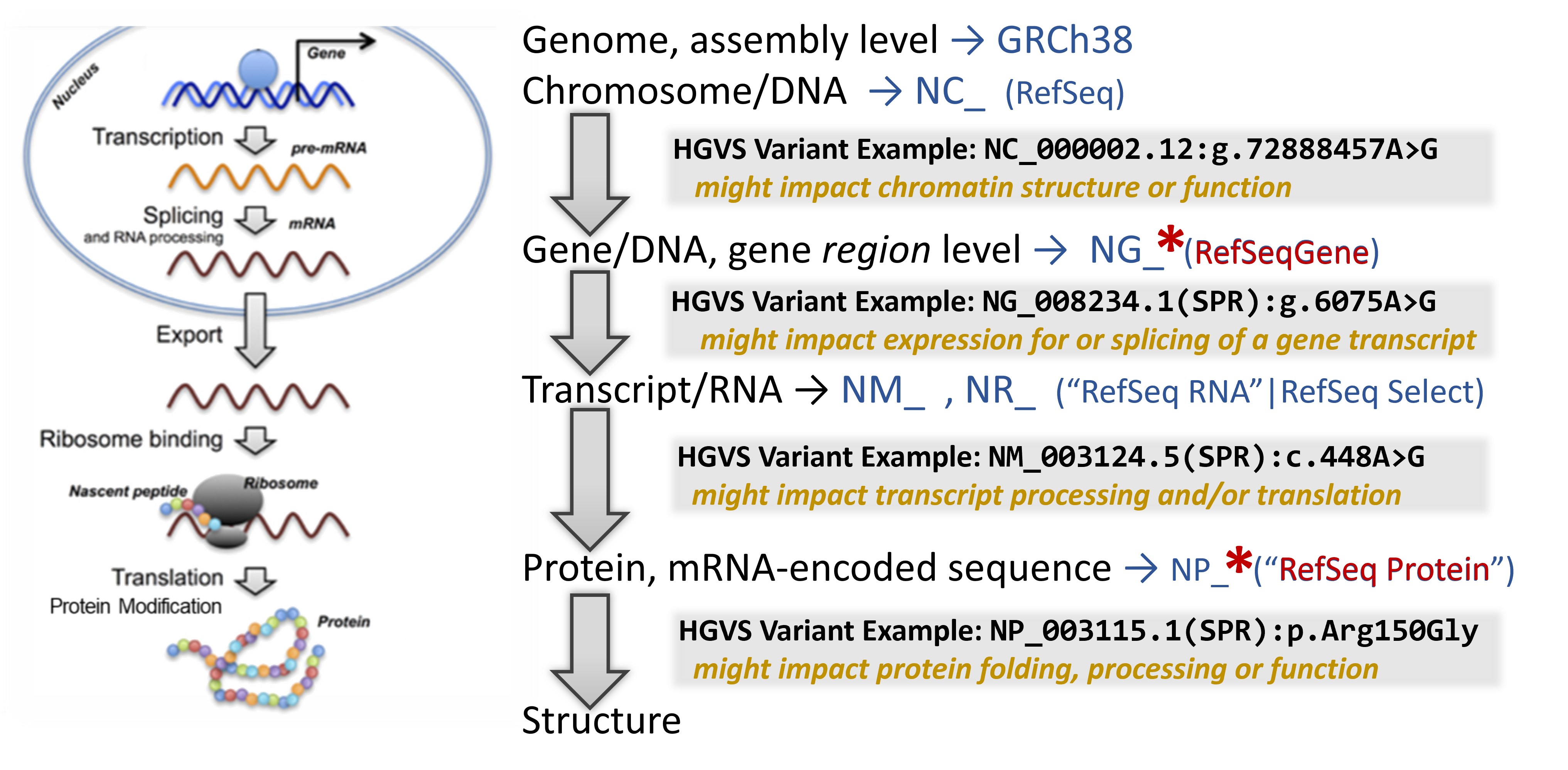
In addition to a lot of helpful aggregated information, NCBI's Gene database provides links to visualization tools which can help to identify where a variant is located in several critical biomolecules.
 If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Notes | |
|
Ultimate Impacted Biomolecule based on:
|
|
-
-
- To learn about the molecular impact of the genetic variant in each boy, begin your search on the relevant NCBI Gene record.
- Click on helpful linked resources to map the location and infer it's impact through this bioinformation flow:
-
 Click here to find a direct link to the GDV display.
Click here to find a direct link to the GDV display.
-
- Note that you are looking at a portion of the chromosome (the accession shown is an NC_).
- Type in the NG_007994.1:g.15392A>G in the top-left text box to search for the variant location.
- Where is the variant located in relation to the indicated gene? (near a gene? upstream or downstream from the gene? in the gene region?)
RefSeqGene Graphic view shows the gene region, including some areas upstream and downstream.
 Click here to find direct links to the RefSeqGene Graphic display.
Click here to find direct links to the RefSeqGene Graphic display.
-
- Type in the NG_007994.1:g.15392A>G in the top-left text box to search for the variant location.
- Where is the variant located in relation to the gene structure?
- Type in the NG_007994.1:g.15392A>G in the top-left text box to search for the variant location.
RefSeq Protein Graphic view shows the protein sequence, including conserved domains and other regions. Look for the record labeled coagulation factor IX isoform 1 preproprotein [Homo sapiens]with the accession that matches what you found in ClinVar: NP_000124.1. (Hint: likely the last one.)
 Click here to find direct links to the RefSeq Protein Graphic display.
Click here to find direct links to the RefSeq Protein Graphic display.
-
- Type in the NP_000124.1:p.Asp110Gly in the top-left text box to search for the variant location.
- Where is the variant located in relation to the protein sequence and it's annotated functional domains?
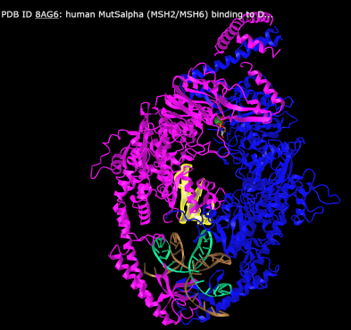
 Click here to see something pretty cool.
Click here to see something pretty cool.
-
- Take a look at the structure, note the annotations for functional regions (conserved domains) and key residues.
- You can check on any particular domain to learn what it's function is.
- Where is the predicted variant-impacted residue in the sequence and in relation to the various annotations in the image?
Let's put it all together to understand what is happening in the patient!
 Bo's parents would like to have some answers.
Bo's parents would like to have some answers.Click here to review some things you may want to consider when formulating the answer to their questions.
-
-
- Which gene is impacted by the genetic variation and what does the gene product normally "do"?
- what is it's biomolecular function?
- what is it's impact on cellular physiology?
- in which cells/tissues is the gene product usually expressed?
- Based on the patient's variation(s):
- what do you think this would do to the gene product's structure and biomolecular function?
- what would this do to cellular physiology?
- what tissues or organs impact be impacted?
- Based on the proposed impacted-tissues/organs, may some of the the patient's symptoms be explained by this? (validating their experience)
- Which gene is impacted by the genetic variation and what does the gene product normally "do"?
-

What do you think is happening in your patient?
| Notes | |
| Diagnosis | |
| Genetic Variation(s) | |
| Proposed Molecular Mechanism of Variant Impact | |
| How does this relate to the phenotype? |
⇑
Take-away message!
Workflow: We've learned a step-by-step process to learn more about a patient's genetic variant that can be used for almost any variant. We've practiced integrating what is learned in each step of the process to help understand the whole picture of molecular biology impacted in the patient - from DNA to gene to transcript to protein and structure. The variant's impact on the protein which is known to be expressed in a particular tissue - can help to explain some of the symptoms that the patient has been experiencing.
Genetic Disorder: Hemophila symptoms can be caused by a couple of different genes. With genetic testing, Bo was found to have a lesion in the F9 gene - identifying the specific disease type of Hemophilia B and identifying the precise gene that should be targeted for therapeutic intervention (either recombinant F9 replacement or Gene Therapy with a functional F9 gene copy).
ANSWER
⇑
PRACTICE CASE with guidance - try this one on your own!
Welcome to Your Patient!
Researching the Referral
-
-
- To learn more about a case, please click on the Referral icon above to open the form.
- Read it over and fill in below what you can glean from the description and proposed preliminary diagnosis.
-

| Notes | |
| Phenotype | |
| Preliminary Diagnosis |
Note: GeneReviews® is a project run by the University of Washington producing expert-authored, point-of-care information with clinically relevant and medically actionable information for inherited conditions. It is an incredible review article-type of resource, and is thus featured in it's own section on relevant MedGen records - as an abstract with links to key sections.
What does the patient's genetic test report say?
In this scenario, the genetic test was selected and performed. The results have come back from the laboratory and have been sent to you for consultation.-
-
- To learn more about Leslie's genetic test results, please click on the Test Results icon above to open the form.
- Read it over and fill in below what you can glean from the report with regard to any genetic variants found and anything they laboratory might be asserting about how this relates to the preliminary diagnosis.
-
 If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Notes | |
| Genetic Variation(s) | |
| Laboratory Assertion(s) |
What is currently known about the identified variant?
Genetic testing laboratories attempt to stay up to date with what is known about the genetic variants that they are assessing. However, it is sometimes valuable to quickly consult with national database of clinical variants (NCBI's ClinVar database) to learn what other organizations have asserted/interpreted for that variant, if anything. In addition to information from testing laboratories, ClinVar receives curated interpretations from authoritative sources such as ClinGen, ACMG and disorder-specific specialist panels. If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Notes on MSH2:g.4951T>C | Notes on MSH2:p.Glu48Ter | |
Variant Information:
|
||
2. Look over the ClinVar records to identify variant's:
- Clinical Interpretation
- Does this match what was listed on the lab report?
- Where on this record can you find out who submitted assertions and what evidence they pointed to?
- HGVS (Human Genome Variation Syntax) aliases, in particular - look for the NG_ and the first NP_ accession-located variants.
- These will be helpful for mapping the variant on these structures and sequences.
- dbSNP Link (and rsID) - The dbSNP database has been collecting biological genetic variant data for 25 years!
- Human population data has been added to relevant variant records from several major projects. What about your patient's variant?
Which of these variants warrant further investigation? Why?
What is currently known about the identified gene?
If a particular gene has been implicated in a genetic test results indicating a pathogenic variant exists in a patient, it is often helpful to understand what that gene is, what its normal function is, where it is found (cellular and tissue expression patterns), and other sources of accessible information, such as links to relevant scientific literature.
NCBI's Gene database aggregates data from many NCBI databases as well as other high-quality resources to provide information and links to help users find and understand what is currently known about a particular organism's gene.
 If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Notes | |
Gene Information on the NCBI Gene record:
|
|
-
-
- To learn more about Leslie's the impacted gene, search NCBI's Gene database with the gene symbol indicated on the genetic test result and find the record for the human version.
- Look over the Gene record that was implicated in the genetic test result and identify:
-
- Summary
- What does this gene normally do? What else has been noted?
- Expression
- In which tissues is this gene normally expressed? (Note there are several tissue sets from different project available to view. Try selecting them in the pull-down menu.)
- Gene Ontology project information
- Where in the cell does this gene product act?
- In what processes/pathways?
- Is there more specific confirmation about what it does?
Does what you've found above make sense based on the patient's symptoms and personal and family history?
Map the variant through the bioinformatic flow!
Now that we understand which gene may be affected by the presence of the detected variant, mapping the variant through the central dogma of molecular biology can help indicate at which point it has its strongest impact.
 Click here to review an overview of the central dogma and genetic variation.
Click here to review an overview of the central dogma and genetic variation.

In addition to a lot of helpful aggregated information, NCBI's Gene database provides links to visualization tools which can help to identify where a variant is located in several critical biomolecules.
 If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Notes | |
|
Ultimate Impacted Biomolecule based on:
|
|
-
-
- To learn about the molecular impact of the genetic variant in each boy, begin your search on the relevant NCBI Gene record.
- Click on helpful linked resources to map the location and infer it's impact through this bioinformation flow:
-
 Click here to find a direct link to the GDV display.
Click here to find a direct link to the GDV display.
-
- Note that you are looking at a portion of the chromosome (the accession shown is an NC_).
- Type in the NP_000242.1:p.Glu48Ter in the top-left text box to search for the variant location.
- Where is the variant located in relation to the indicated gene? (near a gene? upstream or downstream from the gene? in the gene region?)
Based on the location of the variant and the type - what impact do you think it might have on the gene?
 Click here to find direct links to the RefSeqGene Graphic display.
Click here to find direct links to the RefSeqGene Graphic display.
-
- Type in the NG_007110.2:g.5210G>T in the top-left text box to search for the variant location.
- Where is the variant located in relation to the gene structure?
- Type in the NG_007110.2:g.5210G>T in the top-left text box to search for the variant location.
Based on the location of the variant and the type - what impact do you think it might have on the gene expression and resulting transcript?
RefSeq Protein Graphic view shows the protein sequence, including conserved domains and other regions. Look for the record labeled DNA mismatch repair protein Msh2 isoform 1 [Homo sapiens] with the accession that matches what you found in ClinVar: NP_000242.1. (Hint: likely the last one.)
 Click here to find direct links to the RefSeq Protein Graphic display.
Click here to find direct links to the RefSeq Protein Graphic display.
-
- Type in the NP_000242.1:p.Glu48Ter in the top-left text box to search for the variant location.
- Where is the variant located in relation to the protein sequence and it's annotated functional domains?
Based on the location of the variant and the type - what impact do you think it might have on the protein?
Let's put it all together to understand what is happening in the patient!
 Leslie would like to have some answers.
Leslie would like to have some answers.Click here to review some things you may want to consider when formulating the answer to her questions.
-
-
- Which gene is impacted by the genetic variation and what does the gene product normally "do"?
- what is it's biomolecular function?
- what is it's impact on cellular physiology?
- in which cells/tissues is the gene product usually expressed?
- Based on the patient's variation(s):
- what do you think this would do to the gene product's structure and biomolecular function?
- what would this do to cellular physiology?
- what tissues or organs impact be impacted?
- Based on the proposed impacted-tissues/organs, may some of the the patient's symptoms be explained by this? (validating her experience)
- Which gene is impacted by the genetic variation and what does the gene product normally "do"?
-

What do you think is happening in your patient?
| Notes | |
| Diagnosis | |
| Genetic Variation(s) | |
| Proposed Molecular Mechanism of Variant Impact | |
| How does this relate to the phenotype? |
Take-away message!
Workflow: We've practiced this same step-by-step process to learn more about a different patient's genetic variant.
Genetic disorder: Missense variants causing premature termination in the production a critical protein can dramatically compromise both the function of that gene product, but also the function of a key biochemical pathway. In this case, several DNA damage sensing complexes are compromised by the lack of a functional MSH2 protein. With a dysfunctional DNA damage and repair system is not functioning optimally, damage due to intracellular or environmental exposure to mutagens can build up over time and cause dysregulation of cellular growth mechanisms - leading to cancer.
ANSWER
PRACTICE CASES without guidance - try any of these on your own!
| Patient's Name | Patient Photo | Referral & Genetic Test Result Forms | Answers |
| Jeff |  |
||
| Jonathan |  |
|
|
| David |  |
|
Last Reviewed: July 16, 2024