iCn3D Mutation Analysis
Author: Alexa M. Salsbury, PhD
iCn3D Shortcuts and Help Docs
1. Go to 1TUP(MMDB) in iCn3D
Note: you can use your previous rendering or start fresh
2. Click on Select > by Distance
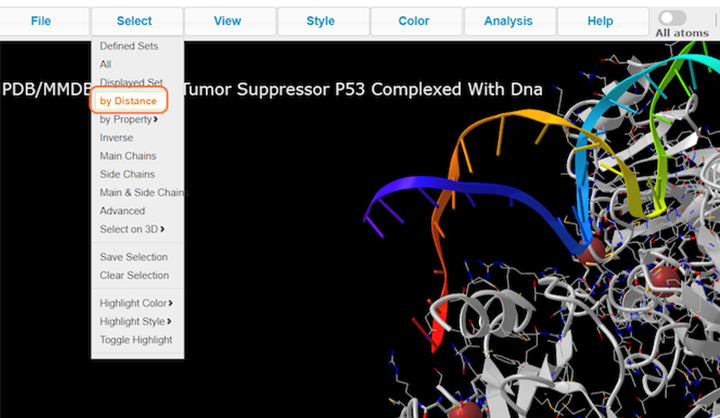
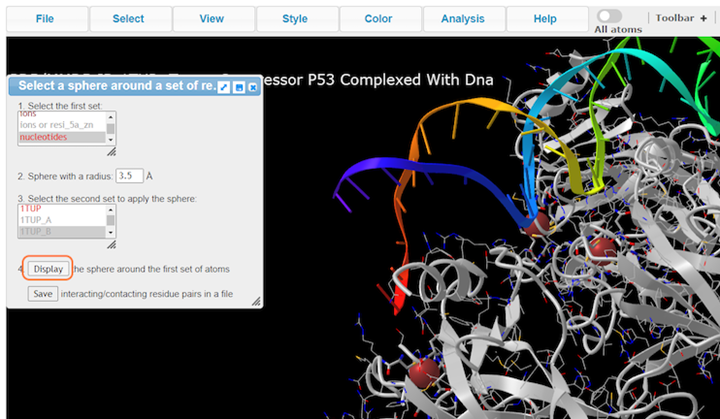
3. Choose nucleotides, set sphere radius to 3.5 Å, choose 1TUP_B, and click on Display
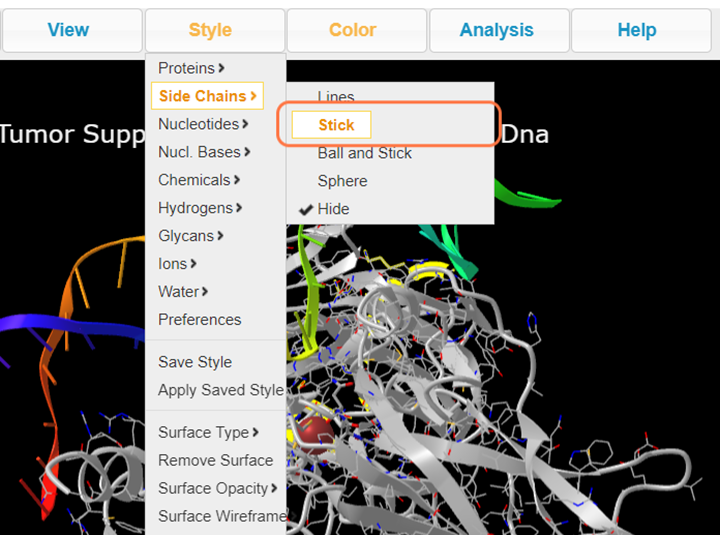
4. Select Style > Side Chains > Stick to better view these residues
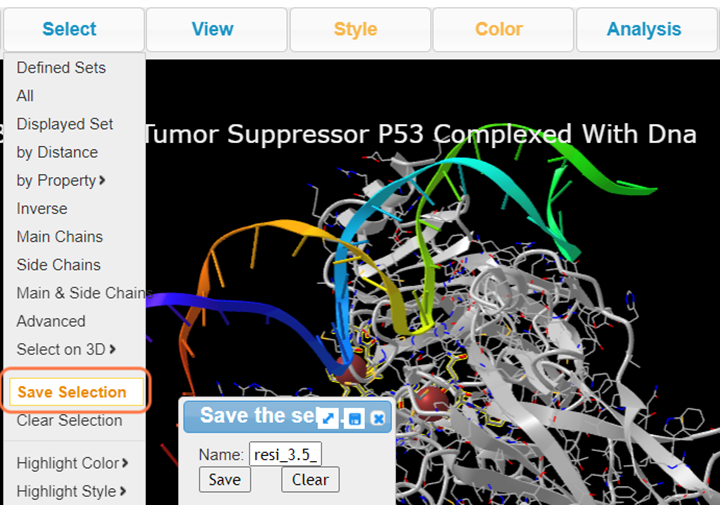
5. Click Select > Save Selection and name resi_3.5_dna
6. In Defined Sets, Select nucleotides +Ctrl resi_5a_zn and View > View Selection to view DNA and the interacting residues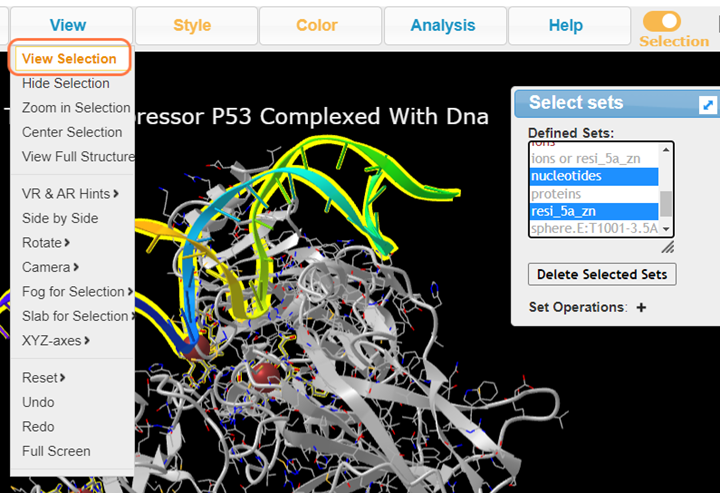
7. View residue Lys120
Note: A handful of residues interacting with the DNA. Lys120 is of particular interest because it’s pointing into the major groove and it’s a positively charged residue.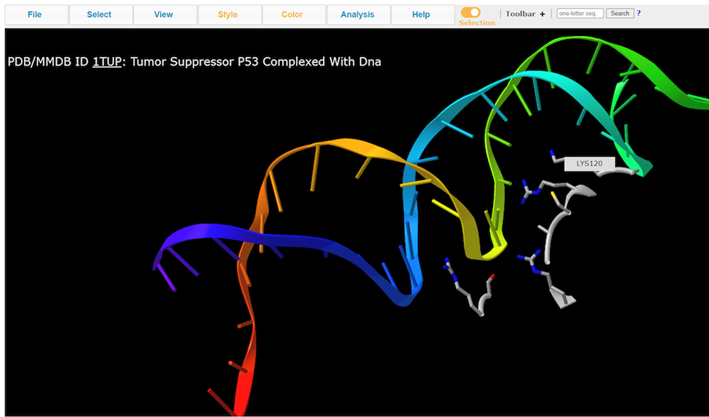
8. Click Analysis > Mutation to perform mutation analysis on Lys120
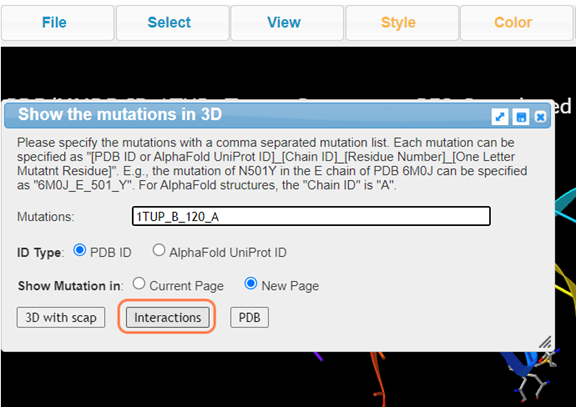
9. Type A to alternate between K120 and A120
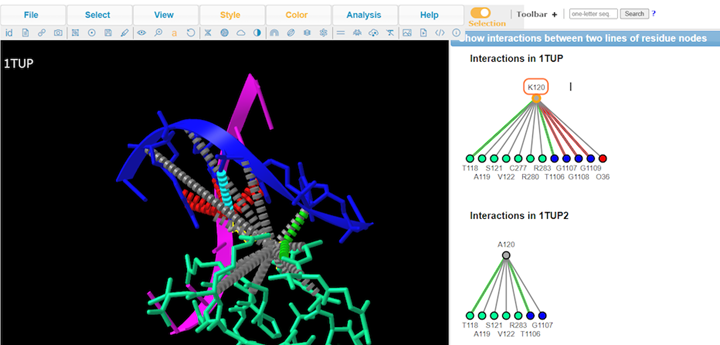
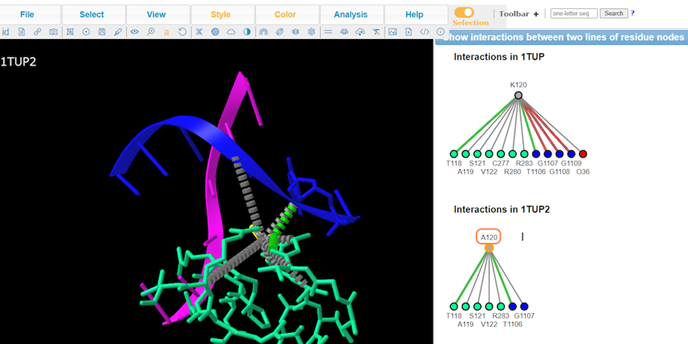
10. Review Key

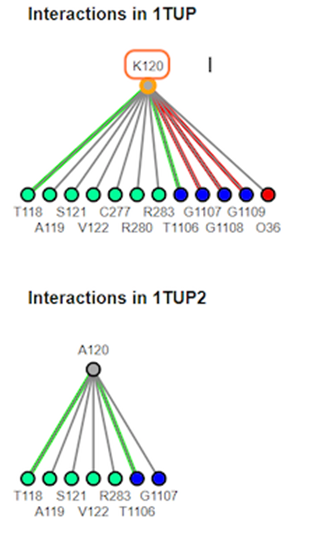
11. Review interaction maps
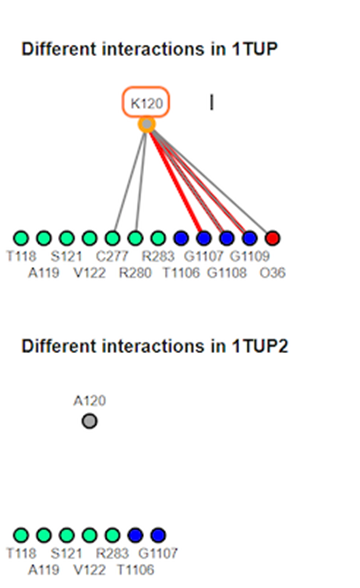
12. Review different interaction maps
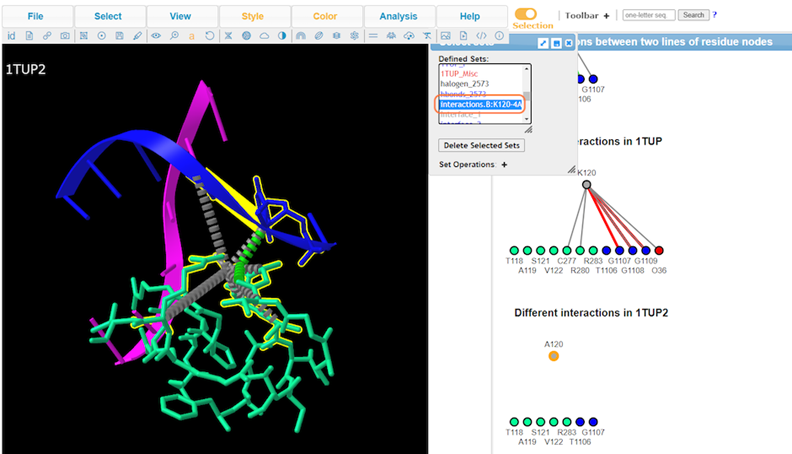
13. View Defined Sets by Analysis > Defined Sets to take a closer look at specific interactions
Page 1 of 1
Last Reviewed: October 18, 2022

