Content
The Polymerase Chain Reaction
The Advent of PCR
In the mid-1980's Kary Mullis and others devised a method to rapidly amplify a targeted region of DNA into milliions of copies. The method used E. coli polymerase mediated extension of DNA oligonucletides (primers) that were complementary to regions flanking the target. They named the method the polymerase chain reaction (PCR). Mullis received the 1993 Nobel Prize in Chemistry for inventing the technique.
Standard PCR uses 30-40 cycles of denaturing a double-stranded target DNA at high temperature, lowering the temperature to allow the primers to bind and the polymerase to extend the primers. Repeated cycles result in an exponential growth of the number of copies of the target DNA.
It's remarkable that the first experiments used a manual process involving water baths and a polymerase that was unstable at high temperatures. The advent of thermostable polymerases from thermophilic bacteria and archaea and computer-controlled thermocycling machines have made PCR a standard laboratory technique that is used in a wide range of molecular biology procedures including measuring gene expression, amplifying targets for sequencing and cloning, and assaying sequence variation.
PCR cycle: denaturation, annealing, extension
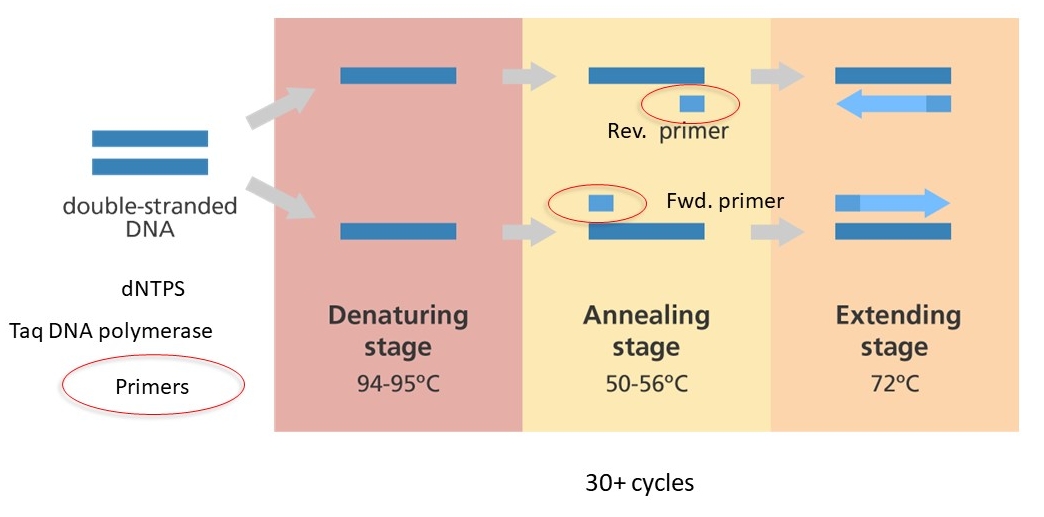
Details of primer binding and extension

Designing primers for PCR
An important aspect of PCR technique is identifying appropriate primers that:
- Match the target sequence in the correct locations
- Have compatible thermodynamic properties with the template such as denaturation temperature (Tm)
- Don't have issues with secondary structure, self-complementarity
- Will not amplify unintended products from the DNA sample
The first three items above are addressed by a primer designing software including the popular and free Primer3. The last item, specificity, requires searching the primers against a database of background DNA sequences to look for unintended matches. This is a job for BLAST.
