iCn3D Mutation Analysis
The objective of this exercise is to better understand interactions within PDB 1TUP, Tumor Suppressor P53 Complexed With DNA, with NCBI's iCn3D. We will use the mutation analysis to probe different mutations discussed in literature (K120A, R248W, and R273H). The embedded links labled "follow" are designed to help participants in the live session follow the instructor's directions. If you fall behind or want to view an instructor created rendering, click these "follow" links.
iCn3D Help Docs
1. Go to iCn3D and load 1TUP
Note: you can use your previous rendering or start fresh
2. Click on Select > by Distance
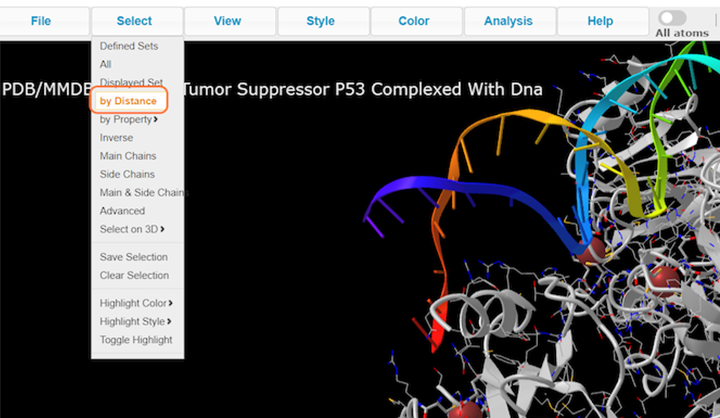
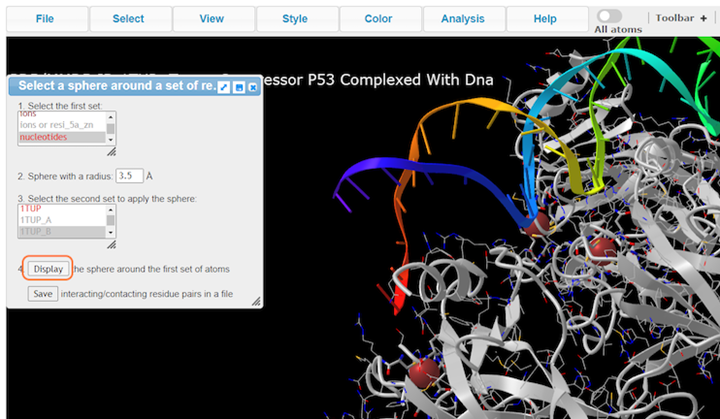
3. Choose nucleotides, set sphere radius to 3.5 Å, choose 1TUP_B, and click on Display
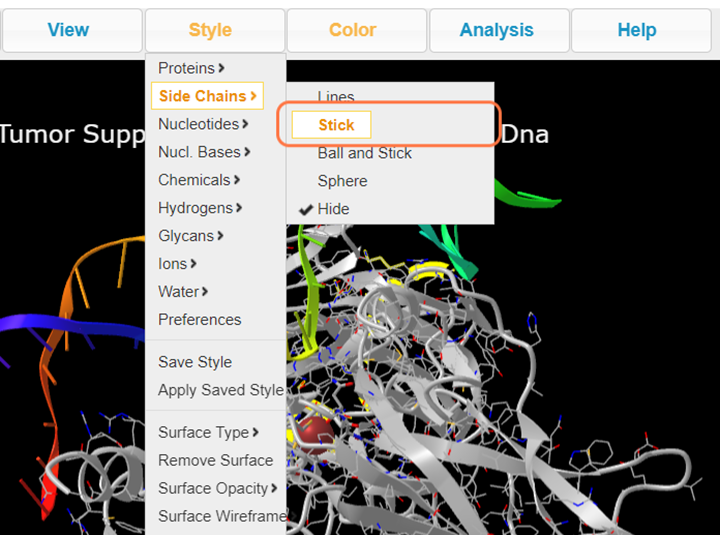
4. Select Style > Side Chains > Stick to better view these residues
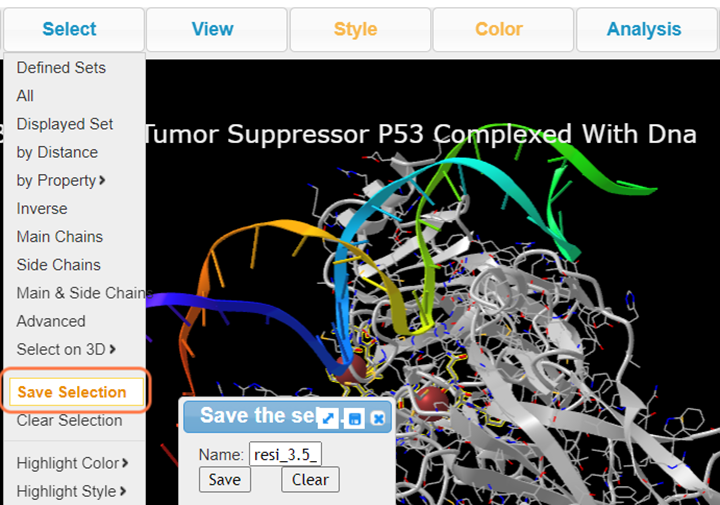
5. Click Select > Save Selection and name resi_3.5_dna
Follow
6. In Defined Sets, Select nucleotides +Ctrl resi_3.5_dna and View > View Selection to view DNA and the interacting residues
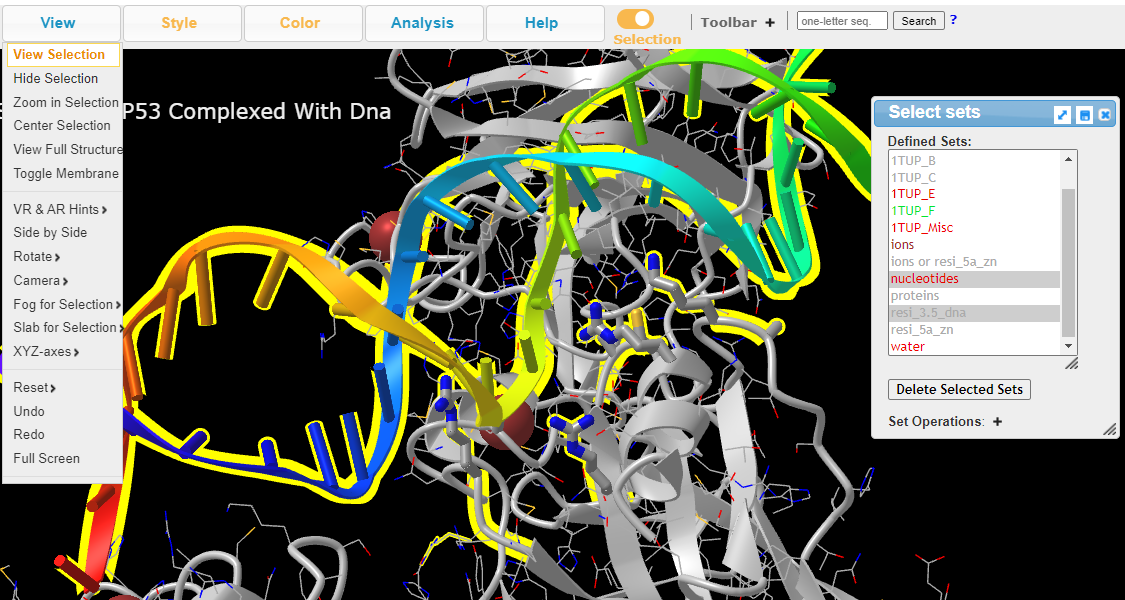
Follow
7. View residue Lys120
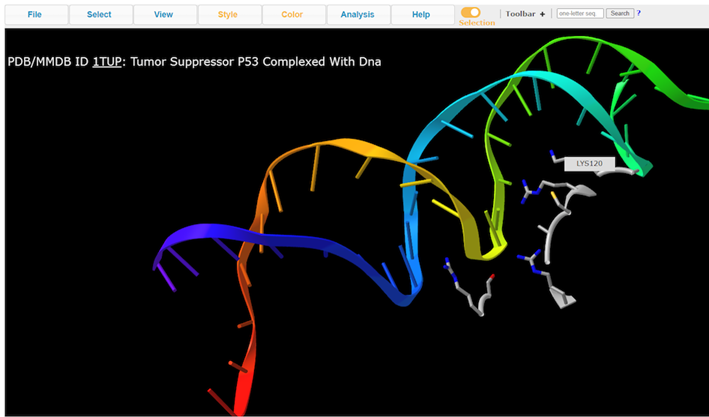
Note: A handful of residues are interacting with the DNA. Lys120 is of particular interest because it’s pointing into the major groove and it’s a positively charged residue.
8. Click Analysis > Mutation and follow the noted format to perform mutation analysis on Lys120
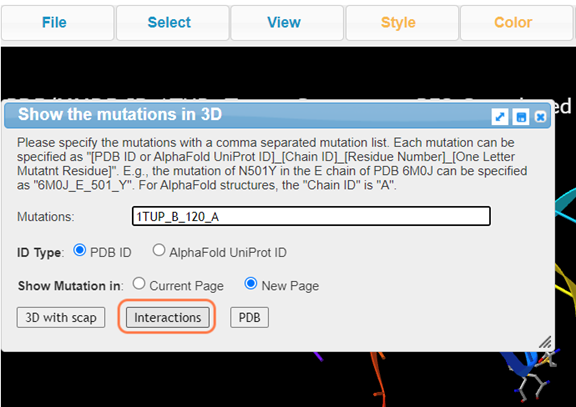
Follow
9. Type A to alternate between K120 and A120
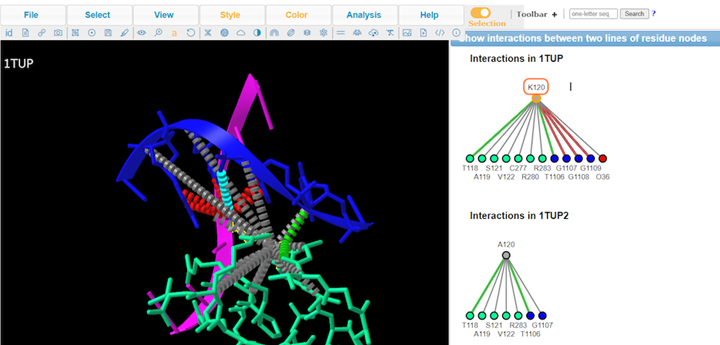
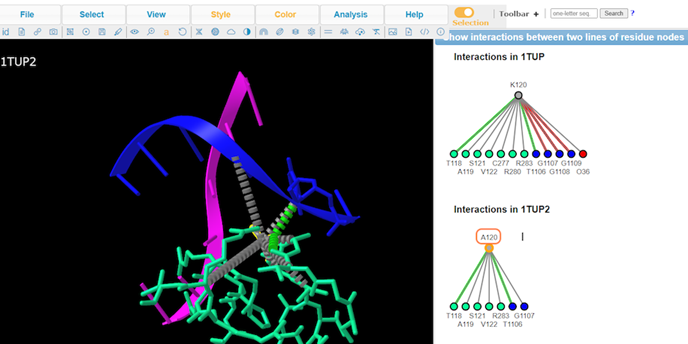
10. Review Key

11. Review interaction maps
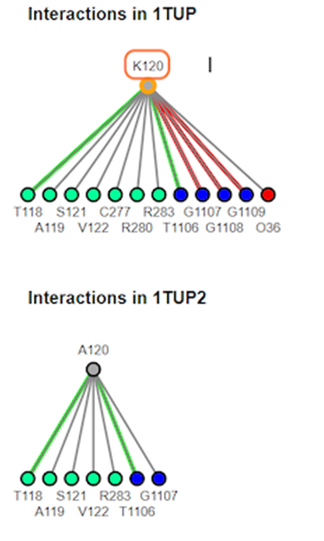
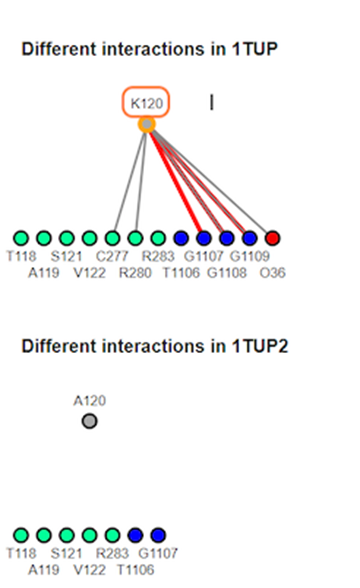
12. Review different interaction maps
13. View Defined Sets by Analysis > Defined Sets to take a closer look at specific interactions
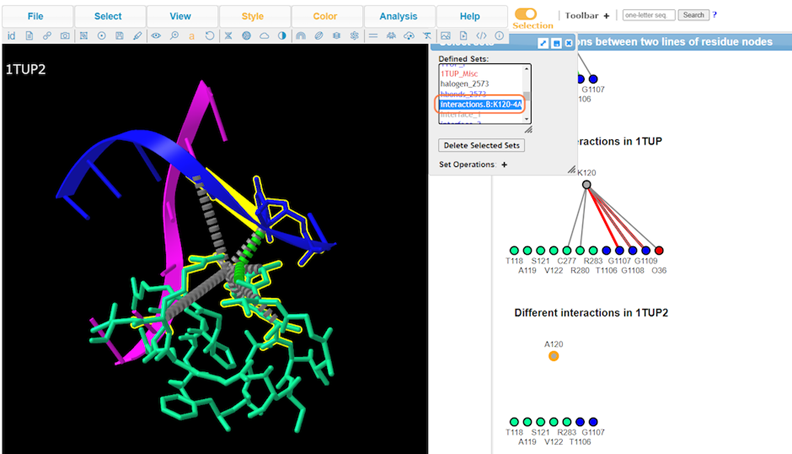
Conclusion:
K120A mutation results in loss of interaction with the DNA groove. The interaction maps generated show that salt bridge/ionic and pie-cation are lost with the substitution.
Practice
Literature shows that Arg248 and Arg273 are common P53 mutations implicated in disease. Use the Mutation analysis to understand how these mutations may affect interactions:
1TUP_B_248_W
1TUP_B_273_H
Conclusion:
These analyses show that R248W mutation results in loss of hydrogen and salt bridge interactions and R273H mutation results in loss of hydrogen bond interactions.
Last Reviewed: February 21, 2023

