Adding Genetics/Genomics to your Clinical Toolkit
How can knowing about a patient's genetic variation help my patient care and case management?
- Validate and specify a diagnosis
- precise variant may diagnose a condition or identify a subtype based on the specific molecular lesion
- may be able to target preventative and/or monitoring efforts even before clinical features become evident
- the specific molecular lesion may help to precisely target a therapy
- Aid to customize and improve a therapeutic outcome
- known variants may explain differences in therapeutic effectiveness due to imact on one or more ADMET issues (absorption, distribution, metabolism, excretion & targeting processes)
- precise variant may point toward information to optimize drug selection and provide dosage prediction
Some vocabulary for clinical genetic testing
- Inherited (germ-line) variants - can be identified in samples from cheek or nasal swab, sputum or blood sample
- Acquired (somatic) variants - would be identified in specific suspect tissue samples (ex: biopsy)
| Currently used diagnosis classifications | Under consideration for additional diagnosis classifications | Currently used pharmacogenetic classifications |
Disorder-causing variants
*An important term to know: |
ACMG: Predisposing variants - Those that increase an individual’s susceptibility to a certain disorder or condition, but where the development of symptoms are not certain (used with the qualifier pathogenic or likely pathognic). ClinGen:
|
Drug response variants
Note: The above were based on CYP450 isoform variants. Some additional terms may be released soon by CPIC. |
-
-
- Variants reported: Specific, pre-designated variants based on a designated clinical phenotype
-
-
-
- Common methods:
- Hybridization/Array
- (increasingly) Next-generation Sequencing (NGS)
- Common methods:
-
-
-
- Variants reported: Laboratories will report on relevant variants if a clinical phenotype code (ICD-10, SnomedCT, HPO ID, OMIM ID) is provided, as is preferred. However, if a clinical code is not specified and one cannot be obtained - all identified with a specific classification (based on the laboratory's policy) may be reported.
- NOTE: Sometimes during NGS analysis a pathogenic or likely pathogenic variant may be detected for a disorder other than indicated by the provided clinical code. This is considered a "Secondary Finding" and may be reported to the ordering clinician. ACMG makes recommendations for reporting these (see below).
- Variants reported: Laboratories will report on relevant variants if a clinical phenotype code (ICD-10, SnomedCT, HPO ID, OMIM ID) is provided, as is preferred. However, if a clinical code is not specified and one cannot be obtained - all identified with a specific classification (based on the laboratory's policy) may be reported.
-
-
-
- Common methods: Next-generation Sequencing (NGS) or Long-read Sequencing
- Single Gene Test (single genome region)
- Multi-gene Panel (several genome regions)
- mDNA test (mitochondrial genome sequence)
- Exome Sequencing (transcript sequence set)
- Whole Genome Sequencing (complete nuclear chromosome set)
- Common methods: Next-generation Sequencing (NGS) or Long-read Sequencing
-
Some key organizations who provide guidance with regard to clinical genetic variants
| Disorder diagnostic variations | ||
| The American College of Medical Genetics and Genomics (ACMG) - supports efforts to improve patient care, establishes standards of care and laboratory policies, and educates members about advances important to their practices. |
ClinGen - an NIH-funded collaborative effort to define the clinical relevance of genes and variants for use in precision medicine and research
|
Disorder-specific organizations often support panels of experts in a particular topic to produce reference information for use by clinical practice groups. |
| Drug selection and dosage variations | ||
| The Clinical Pharmcogenetics Implementation Consortium (CPIC) - curates and posts, peer-reviewed, evidence-based, updatable, and detailed gene/drug clinical practice guidelines. |
The Pharmacogenomics Knowledgebase (PharmGKB) - a resource that aggregates Pharmacogenomics Research Network (PGRN)-curated information about the impact of genetic variation on drug response for clinicians and researchers. |
The U.S. Food and Drug Administration (FDA) - maintains a table of drug-gene interactions with sufficient scientific evidence to suggest that subgroups of patients may be likely to have altered drug metabolism. FDA approved guidelines for incorporating genetics in selection or dosage for a specific medication are listed on relevant drug labels.
|
What does this have to do with NCBI?
We are a “center” within the NLM responsible for creation, curation and maintenance of medical and scientific databases and other things…![]()
We receive, create, archive & make available biomedical information, as well as perform computational biology & IT systems research… 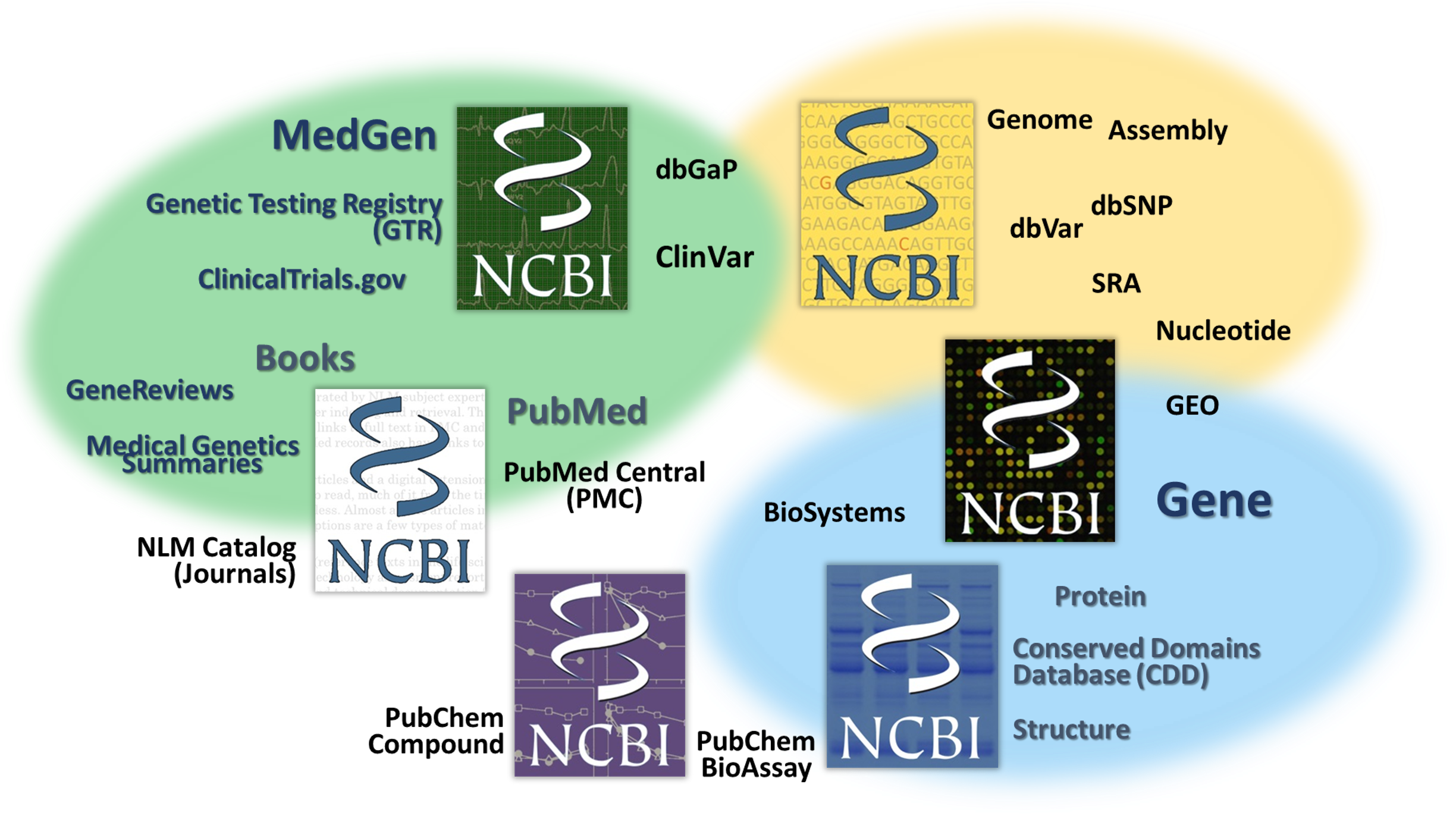
We really aspire to help make sense and promote good use of the information!
If you are looking for information on gene-associated disorders, genetic tests and what is known about genetic variations......
Here are some key NCBI resources where you can START to learn!
Last Reviewed: April 17, 2023

