Learning the workflow with four real cases
The cases
 |
 |
 |
 |
What do we know about the patient's phenotype?
In this scenario, each of four boys was referred to the genetics clinic because their preliminary diagnoses suggested they might have a particular condition impacted by potentially pathogenic genetic variation(s). If a known pathogenic genetic variant is found, it can validate the diagnosis and confirm a specific subtype for that disorder.
-
-
- To learn more about a case, please click on the Referral icon under the name.
- Read it over and fill in below what you can glean from the description and proposed preliminary diagnosis.
- As a group, we'll compare your patient's information with what others find for the other cases.
-
| Marco | Alexei | James | Bo | |
| Phenotype (including severity) | ||||
| Preliminary Diagnosis |
What does the patient's genetic test report say?
In this scenario, the genetic test was selected and performed. The results have come back from the laboratory and have been sent to you for consultation. If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Marco | Alexei | James | Bo | |
| Genetic Variation(s) | ||||
| Laboratory Assertion(s) |
-
-
- To learn more about your patient's genetic test results, please click on the Test Results icon under his name
- Read it over and fill in below what you can glean from the report with regard to any genetic variants found and anything they laboratory might be asserting about how this relates to the preliminary diagnosis.
- As a group, we'll compare your patient's information with what others find for the other cases.
-
What is currently known about the identified variant?
Genetic testing laboratories attempt to stay up to date with what is known about the genetic variants that they are assessing. However, it is sometimes valuable to quickly consult with national database of clinical variants (NCBI's ClinVar database) to learn what other organizations have asserted/interpreted for that variant, if anything. In addition to information from testing laboratories, ClinVar receives curated interpretations from authoritative sources such as ClinGen, ACMG and disorder-specific specialist panels. If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
 |
 |
 |
 |
|
Variant Information:
|
||||
-
-
- To learn more about your patient's genetic variant(s), search NCBI's ClinVar database with the variant indicated on the genetic test results.
- Look over the ClinVar record for the variant and identify:
-
- Clinical Interpretation
- Does this match what was listed on the lab report?
- Where on this record can you find out who submitted assertions and what evidence they pointed to?
- HGVS (Human Genome Variation Syntax) aliases, in particular - look for the NG_ and the first NP_ accession-located variants.
- These will be helpful for mapping the variant on these structures and sequences.
- dbSNP Link (and rsID) - The dbSNP database has been collecting biological genetic variant data for 25 years!
- Human population data has been added to relevant variant records from several major projects. What about your patient's variant?
What is currently known about the identified gene?
If a particular gene has been implicated in a genetic test results indicating a pathogenic variant exists in a patient, it is often helpful to understand what that gene is, what its normal function is, where it is found (cellular and tissue expression patterns), and other sources of accessible information, such as links to relevant scientific literature.NCBI's Gene database aggregates data from many NCBI databases as well as other high-quality resources to provide information and links to help users find and understand what is currently known about a particular organism's gene.
 If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
 |
 |
 |
 |
|
Gene Information on the NCBI Gene record:
|
||||
-
-
- To learn more about the impacted gene in each boy, search NCBI's Gene database with the gene symbol indicated on the genetic test result and find the record for the human version.
- Look over the Gene record for the variant and identify:
-
- Summary
- What does this gene normally do? What else has been noted?
- Expression
- In which tissues is this gene normally expressed? (Note there are several tissue sets from different project available to view. Try selecting them in the pull-down menu.)
- Gene Ontology project information
- Where in the cell does this gene product act?
- In what processes/pathways?
- What does the gene product actually do?
Map the variant through the bioinformatic flow!
Now that we understand which gene may be affected by the presence of the detected variant, mapping the variant through the central dogma of molecular biology can help indicate at which point it has its strongest impact.Note: Currently, genetic testing laboratories most often use genomic DNA for their genetic tests - identifying a variant by its position on a particular chromosome. They most often report variants found in gene regions, since currently this is where most research has been focused on to explain impacts on human biology.
 Click here to review an overview of the central dogma and genetic variation.
Click here to review an overview of the central dogma and genetic variation.
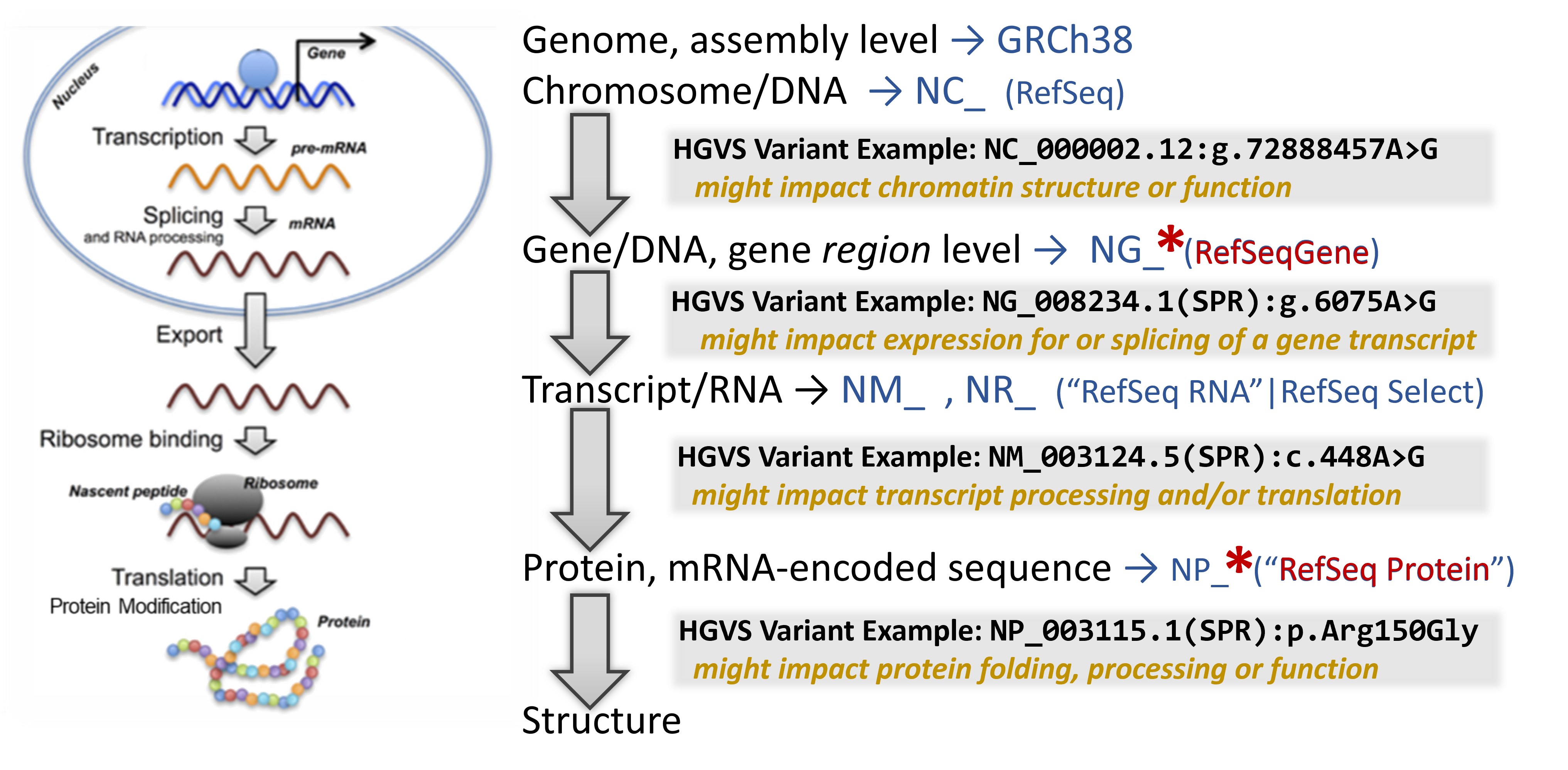
In addition to a lot of helpful aggregated information, NCBI's Gene database provides links to visualization tools which can help to identify where a variant is located in several critical biomolecules.
 If you want it, click here to display the portion of the form for this section.
If you want it, click here to display the portion of the form for this section.
| Marco | Alexei | James | Bo | |
|
Ultimate Impacted Biomolecule based on:
|
||||
-
-
- To learn about the molecular impact of the genetic variant in each boy, begin your search on the relevant NCBI Gene record.
- Click on helpful linked resources to map the location and infer it's impact through this bioinformation flow:
-
Genome Data Viewer shows the chromosome zoomed into the gene region.
-
- Note that you are looking at a portion of the chromosome (the accession shown is an NC_).
- Type in the NG_ HGVS in the top-left text box to search for the variant location.
- Find where the variant is located in relation to the indicated gene.
RefSeqGene Graphic view shows the gene region, including some areas upstream and downstream.
-
- Type in the NG_ HGVS in the top-left text box to search for the variant location.
- Find where is the variant located in relation to the gene structure.
- Type in the NG_ HGVS in the top-left text box to search for the variant location.
RefSeq Protein Graphic view shows the protein sequence, including conserved domains and other regions.
-
- Type in the NP_ HGVS in the top-left text box to search for the variant location.
- Find where is the variant located in relation to the protein sequence and it's annotated functional domains.
- Type in the NP_ HGVS in the top-left text box to search for the variant location.
From here you might click to see see the protein's Conserved Domains or visualize the 3D structure.
-
- Take a look at the structure, note the annotations for functional regions (conserved domains) and key residues.
- You can check on any particular domain to learn what it's function is.
Let's put it all together to understand what is happening in the patient!
The boys' parents would like to have some answers.
Click here to review some things you may want to consider when formulating the answer to her questions.
- Which gene is impacted by the genetic variation and what does the gene product normally "do"?
- what is it's biomolecular function?
- what is it's impact on cellular physiology?
- in which cells/tissues is the gene product usually expressed?
- Based on the patient's variation(s):
- what do you think this would do to the gene product's structure and biomolecular function?
- what would this do to cellular physiology?
- what tissues or organs impact be impacted?
- Based on the proposed impacted-tissues/organs, may some of the the patient's symptoms be explained by this? (validating the patient's experience)
 What do you think is wrong and happening with your patient?
What do you think is wrong and happening with your patient?
 |
 |
 |
 |
|
| Diagnosis | ||||
| Genetic Variation(s) | ||||
| Proposed Molecular Mechanism of Variant Impact | ||||
| How does this relate to the phenotype? |
Take-away message!
Workflow: We've learned a step-by-step process to learn more about a patient's genetic variant that can be used for almost any variant.
Genetic disorders: Predicting the likelihood or timing of onset and progression of a genetic disorder based on a patient's genetic variant(s) is not as simple as hoped for.
- Different genetic/molecular mechanisms may produce similar phenotypes/disorders - however, the proper selection of targeted therapies often requires knowledge of the precise molecular lesion.
- For example, recombinant replacement or gene therapy for Hemophilia requires targeting the specific pathological mechanism - malfunction of a specific blood clotting factor.
- Patients with the same affected gene may experience different levels of symptom severity or even diagnosable conditions.
- For example, Bo's bleeding issues were less severe than Alexei's.
- Of note: There are vastly different presenting genetic disorders that are caused by different genetic variations in the same gene. For example: LMNA is associated with Hutchinson-Gilford progeria, Emery-Dreifuss muscular dystrophy, Familial lipodystrophy, and even a specific type of dilated cardiomyopathy.
ANSWERS
Last Reviewed: May 23, 2023

