Research Tips for Genetic Disorder & Variant Mapping
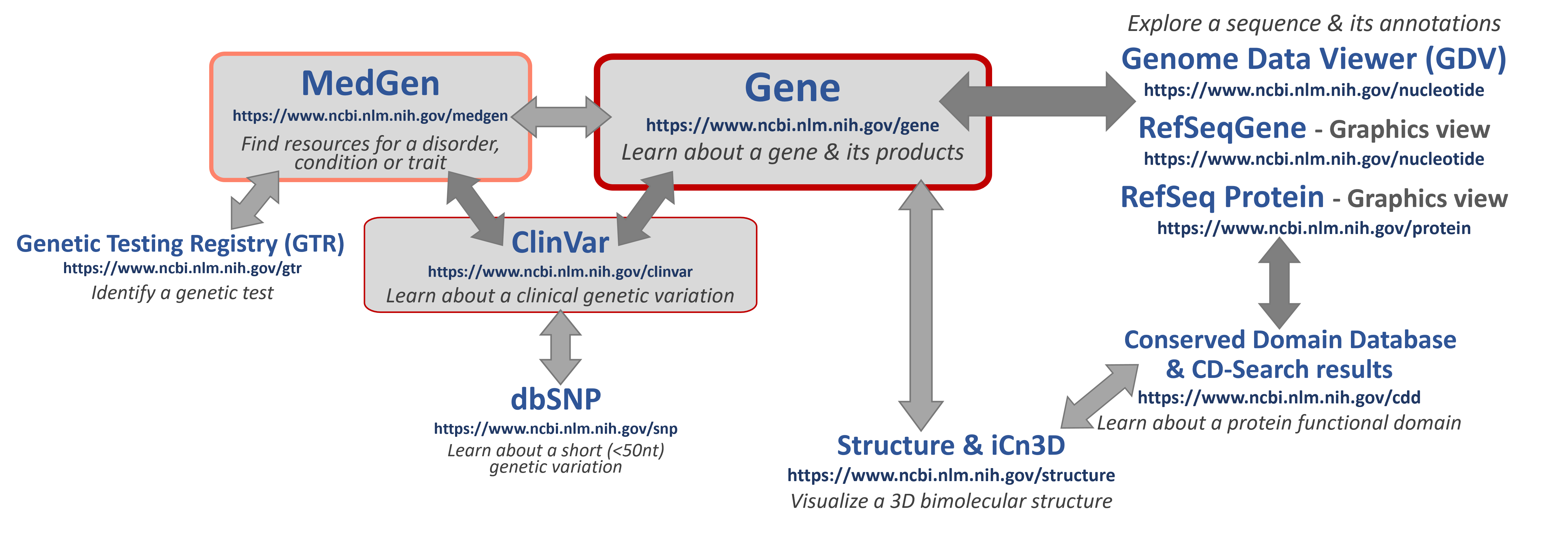
Available Resources at NCBI
If you are looking for information here are some good places to START to learn about key topics!
Here are some helpful database hubs for human variation research:
Here are some good tools for mapping variants on to biomolecular structures:
- MedGen - an aggregate database for information and links to more about gene-associated human disorders and phenotypes.
- Genetic Testing Registry (GTR) - a source for clinical and research genetic tests with information provided by the laboratories.
- ClinVar - a genetic variation resource collating clinically-relevant information submitted by research and clinical labs, expert clinical panels, and some key genetic disease literature resources.
- dbSNP - a registry of short human genetic variants with biological information and population data.
- NCBI Gene - an aggregate database for information and links to more data about genes
- NCBI Structure - a source for curated 3D biomolecular structure information based on submissions to the Protein DataBank (PDB).
Here are some good tools for mapping variants on to biomolecular structures:
-
BLAST - genome or reference sequence databases
-
Genome Data Viewer (GDV) - a full-service interactive genome sequence and annotation browser
-
On NCBI Nucleotide or NCBI Protein database record:
- Graphical Sequence Viewer - an interactive sequence browser display available by clicking on "Graphics"
- Pre-calculated Conserved Domain (CD) View - an interactive graphical display of CD-Search results by clicking on "Identify Conserved Domains"
-
iCn3D - a web-based 3D structure viewer accessible on NCBI Structure record pages or as a stand-alone tool.
Variant nomenclature
Over the years, researchers have adopted many different ways to name a particular genetic variant that they have been studying. Here are some examples of what has been used in published literature for exactly the same genetic variant:
-
-
- Factor V Leiden variant
- F5 Arg534Gln
- FV R506Q
- NC_000001.10:g.169519049=
- NC_000001.11:g.169549811C>T
- rs6025
- OMIM: 612309.0001
-
A standard way to notate these has been proposed - the Human Genome Variant Syntax (HGVS) and is now often in use by many research and clinical labs. This notation is based on anchoring the location of a variant to a specific, discrete sequence record.
Accession.version(gene symbol):molecular type-abbreviation.
then a structured statement including:
variant-location, wildtype-residue and variant-impact (such as the variant residue)
then a structured statement including:
variant-location, wildtype-residue and variant-impact (such as the variant residue)
For example:
NG_011806.1(F5):g.41721G>A
or
NP_000121.2(F5):p.Arg534Gln
For Accession.version, a particular set of sequences and their accessions is often used to reliably and sustainably anchor these variants. The NCBI RefSeq Project collects all known nucleotide and protein sequences and uses them and literature information to create a non-redundant set of reference sequences (recognizable accession prefixes). For humans, these are the most commonly found record-types and prefixes for their accessions:
-
- Chromosome: NC_
- Gene/Gene region: NG_
- Transcript
- Protein coding: NM_ (with strong evidence) or XM_ (predicted)
- Non-protein coding: NR_ (with strong evidence) or XR_ (predicted)
- Protein with sequence translated from the transcript: NP_ (with strong evidence) or XP_ (predicted)
Based on usage in the community, the inclusion of a gene symbol, though recommended, appears to be optional. However if included, it should be the official gene symbol as designated by the Human Genome Nomenclature Committee (HGNC).
For molecular type-abbreviation, at NCBI you will often see:
-
- “g.” for a linear genomic reference sequence
- “c.” for a coding DNA reference sequence
- “p.” for a protein reference sequence
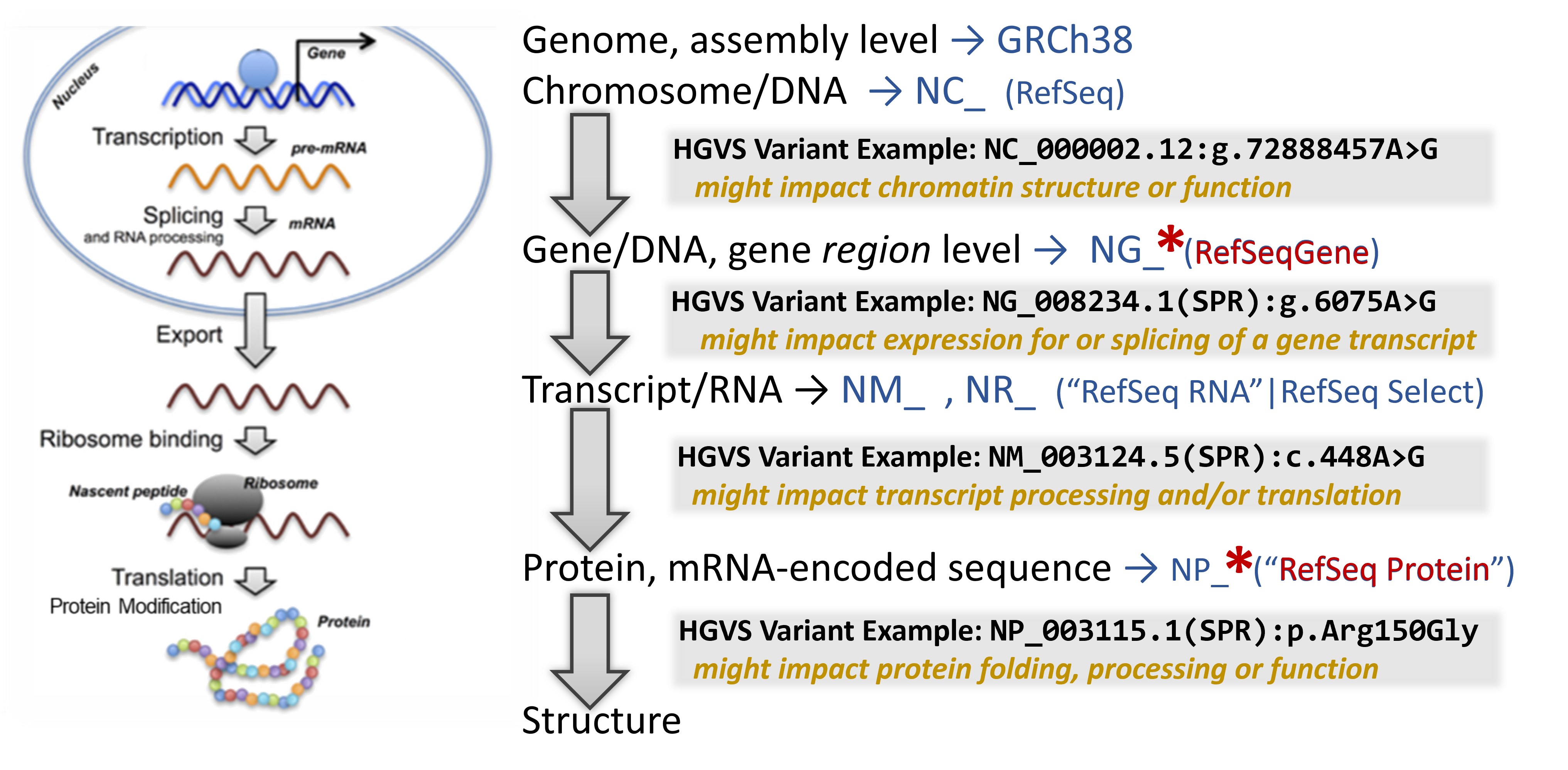
For examples of HGVS usage, take a look below in the overview of how these fit in with biomolecules throughout the central dogma of molecular biology.
A quick primer on the central dogma & how genetic variants can impact molecular biology
Let's put this all together in a general workflow that you can use
Last Reviewed: May 20, 2023