Explore On Your Own

Use this time to:
- Try out your favorite organisms
- Work through three scenarios, plus a Skills Test in Scenario #3
- Ask questions
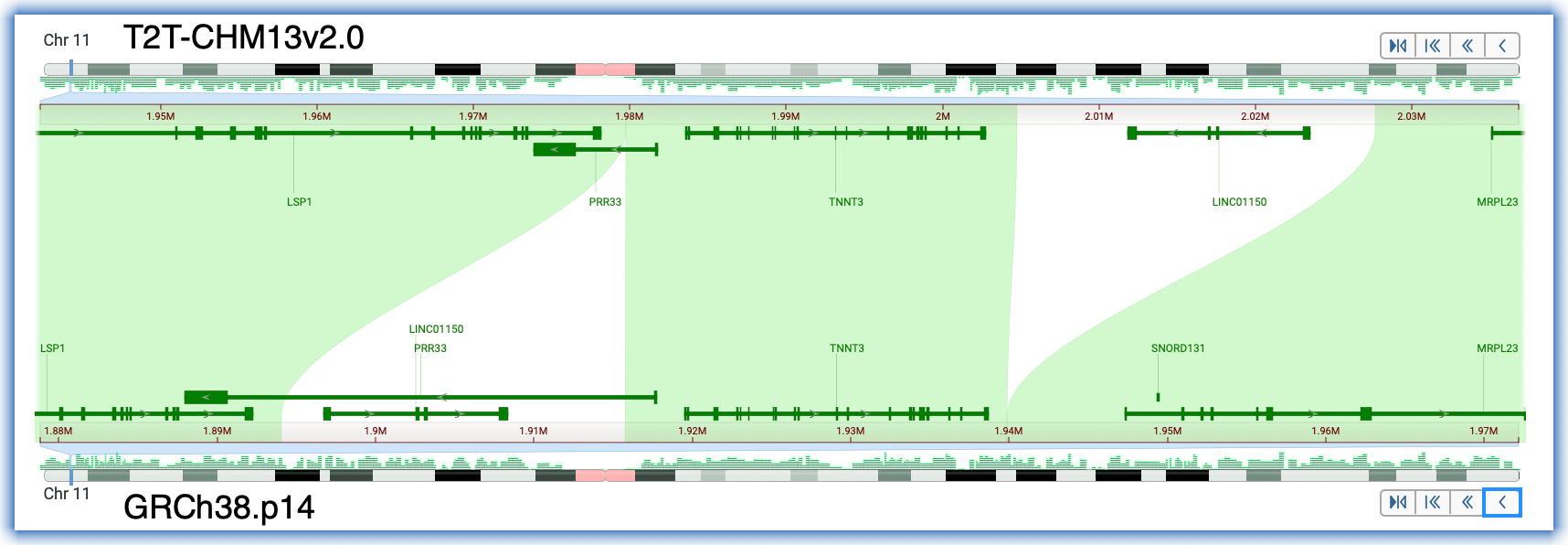
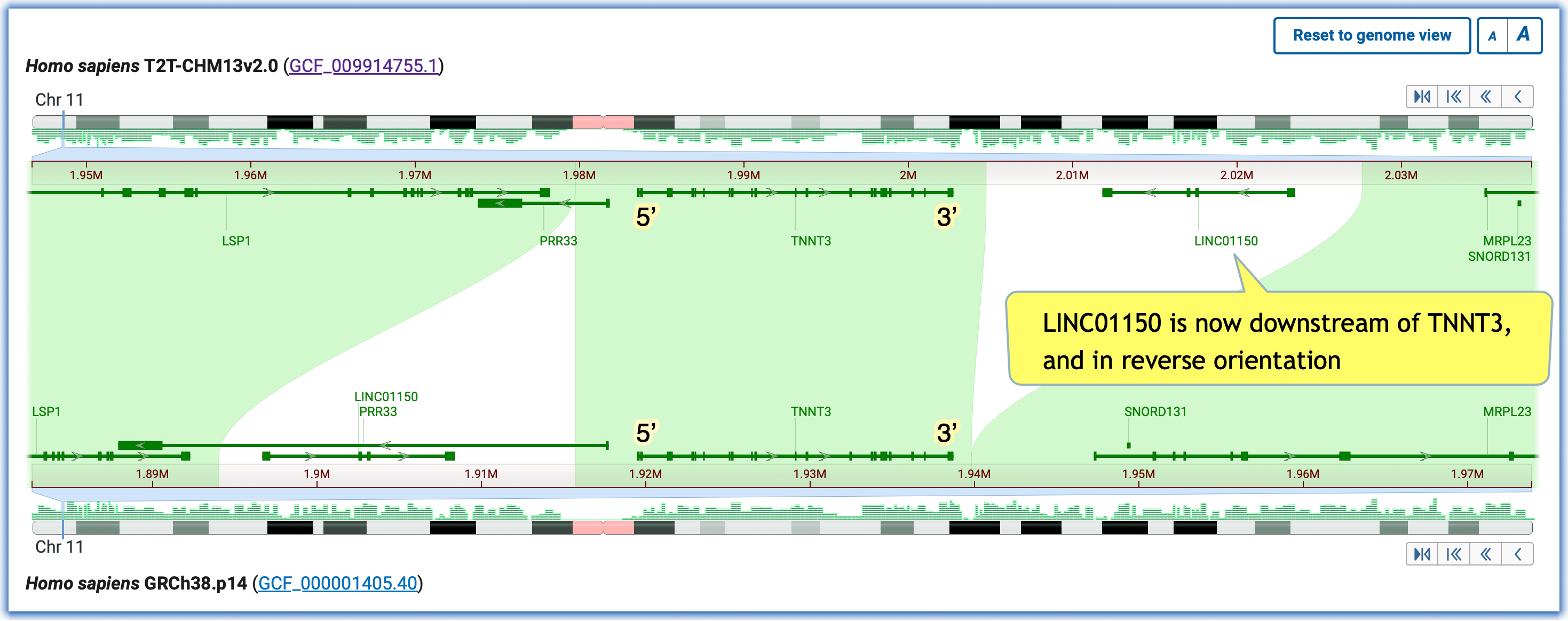
Scenario #1: Rearrangement of a 22kb region upstream of the human TNNT3 gene, which is implicated in forms of arthrogryposis.
Click here for the workflow exploring the TNNT3 rearrangement
- Set up the alignment for human T2T-CHM13v.2.0 and human GRCh38.p14
- Search for the gene symbol, TNNT3
- Click in the Description row for either assembly
- Adjust the zoom to see the extent of the unaligned regions on both sides of TNNT3
- Notice the orientation of TNNT3, the changed orientation of LINC01150, and closer proximity to its genetically-linked partner, TNNI2.
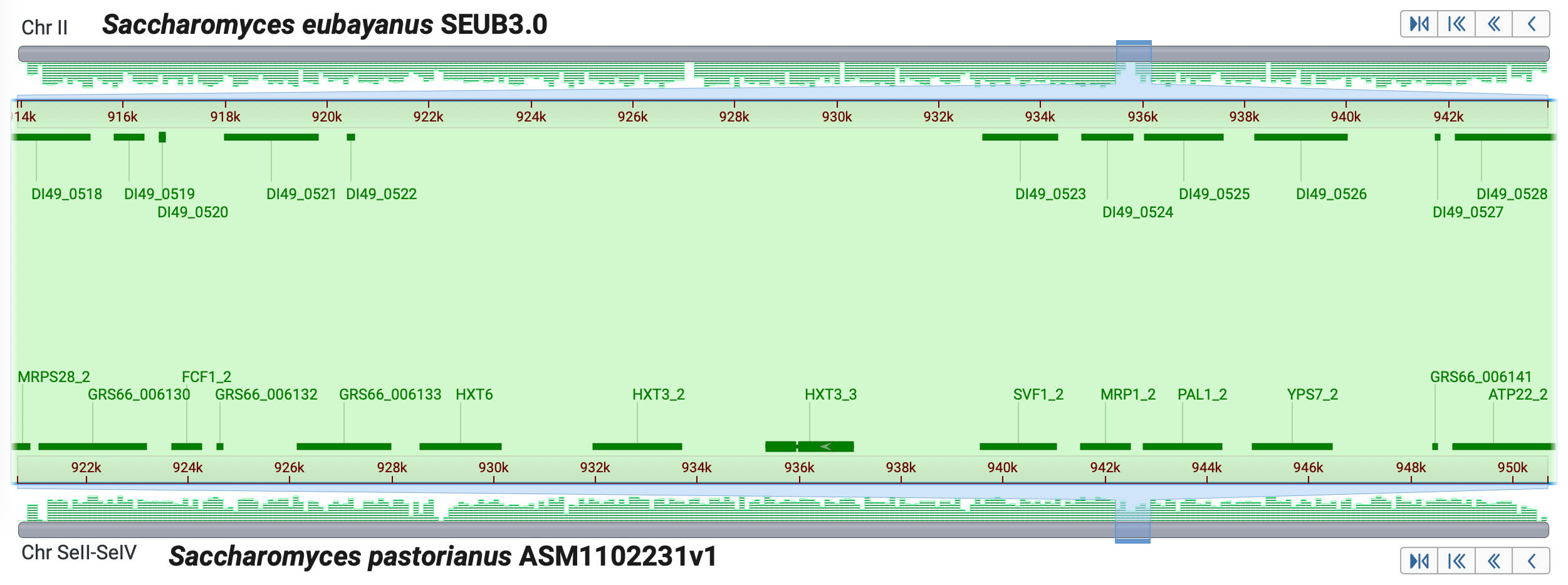
Scenario #2: Compare two yeast species used in fermentation to explore the hexose transporter, HXT, genes. Below is Saccharomyces eubayanus and Saccharomyces pastorianus
Click here for the workflow exploring the HXT genes
- Set up the alignment for Saccharomyces eubayanus and Saccharomyces pastorianus
- Search for the gene symbol, hxt3
- Click in the Description row for DI49_0522, assembly SEUB3.0 (S. eubaynus)
- Adjust the zoom as in the image below, guided by the gene order synteny
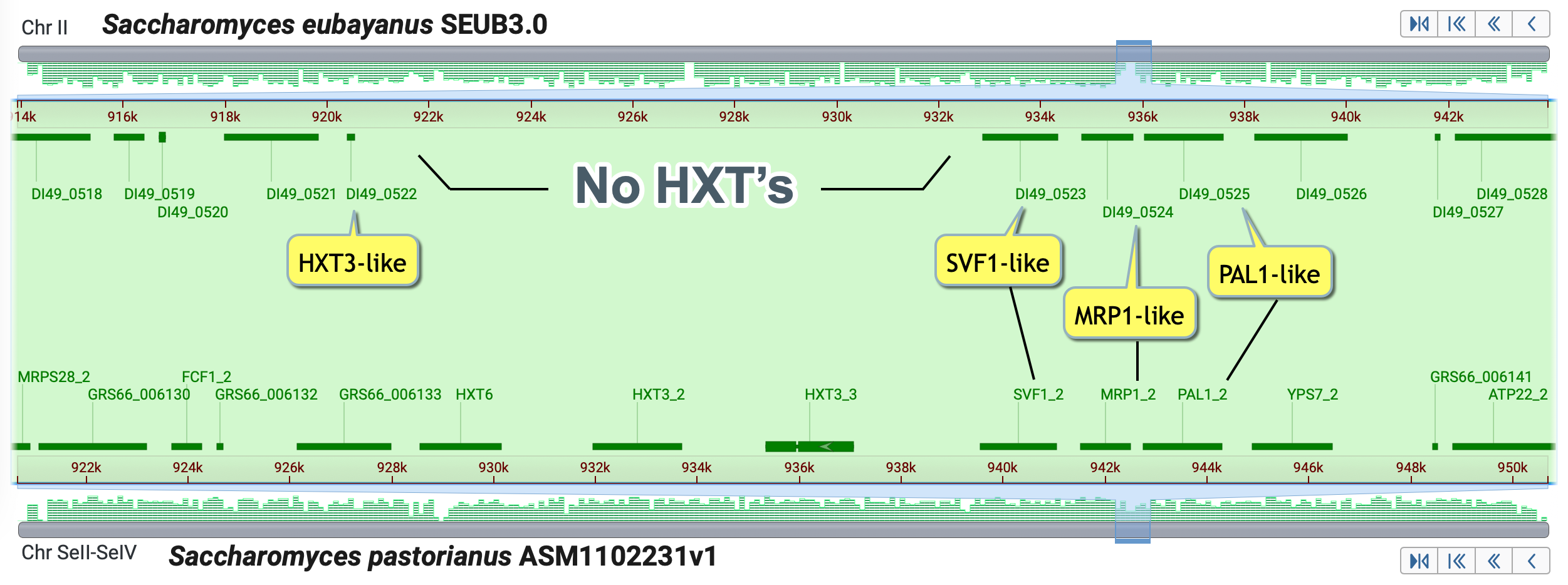
- Notice the lack of HXT genes in this region of SEUB3.0
Optional Exercise
What are possible explanations for the paucity of HXT genes in this region of S. eubayanus?Use the MSA Viewer and/or the GDV to explore the region.
Click here to see how the MSA presents one possible explanation
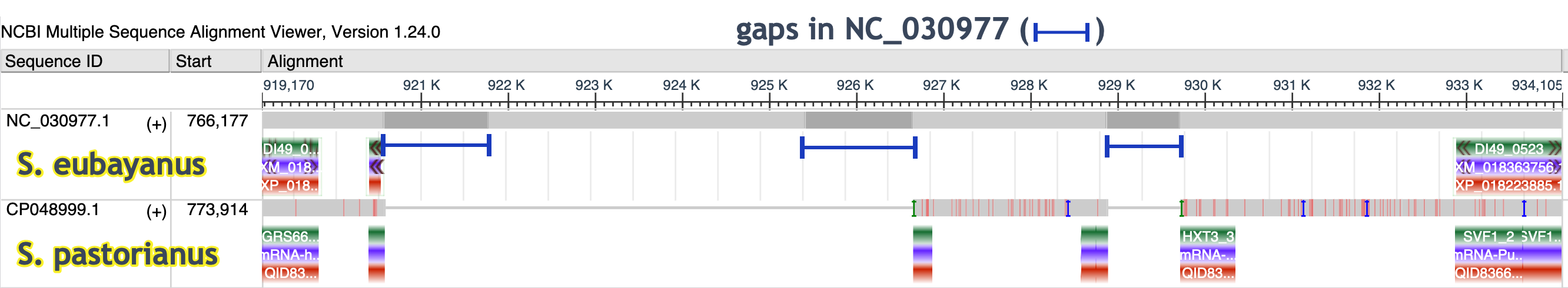
The gaps in the assembled chromosome record, NC_030977.1, complicate annotation in this region.
Scenario #3: Interesting comparison: the tetraploid African clawed frog, Xenopus laevis, and the diploid tropical clawed frog, Xenopus tropicalis.
From: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10158840/
"Xenopus laevis is an allotetraploid due to an ancient hybridization event resulting in 1 set of 9 long (L) chromosomes and a second set of 9 short (S) chromosomes, so there are 2 X. laevis genes for each locus (with some exceptions), while there is a single gene for each locus found in X. tropicalis"
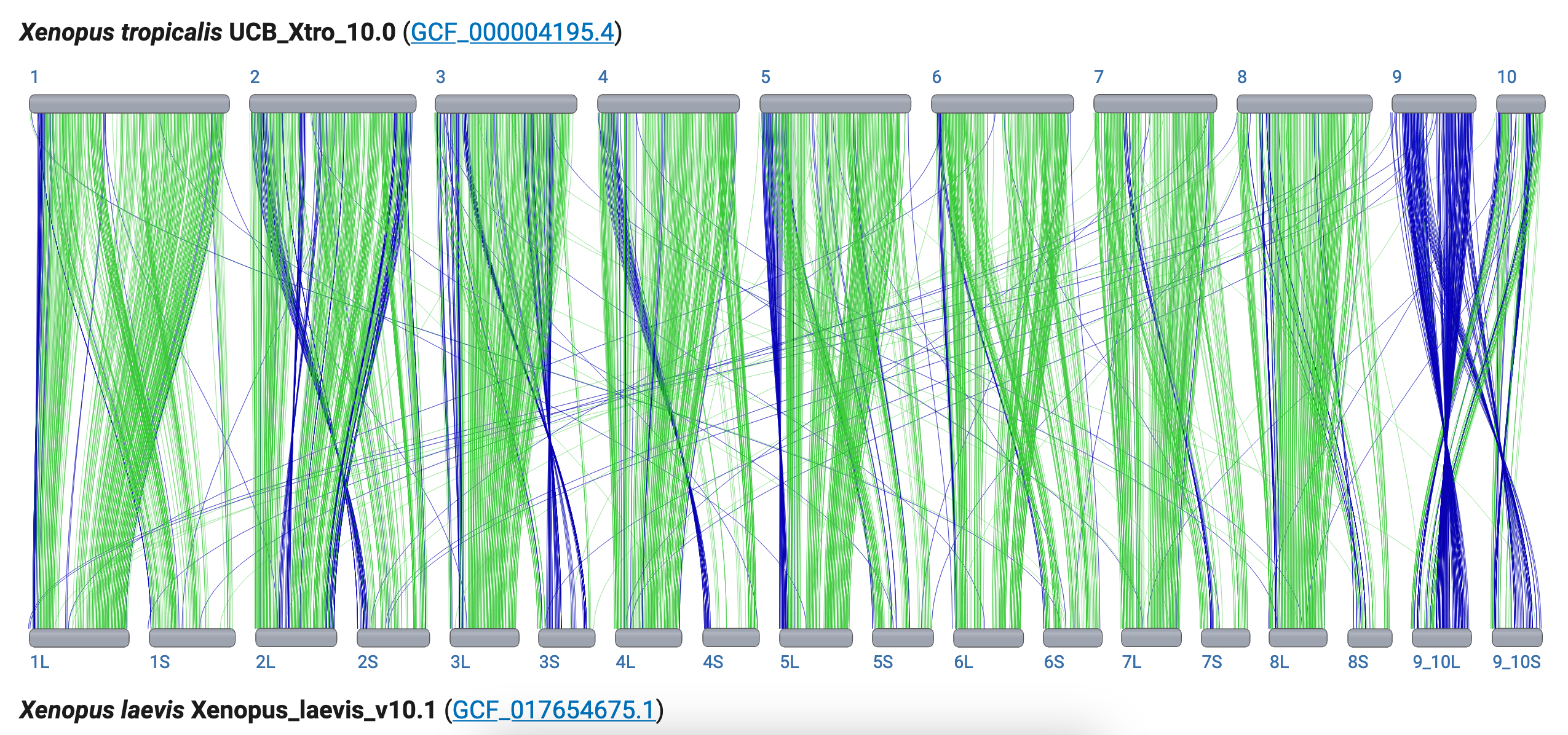
Use the X. tropicalis / X. laevis comparison to test your CGV skills.
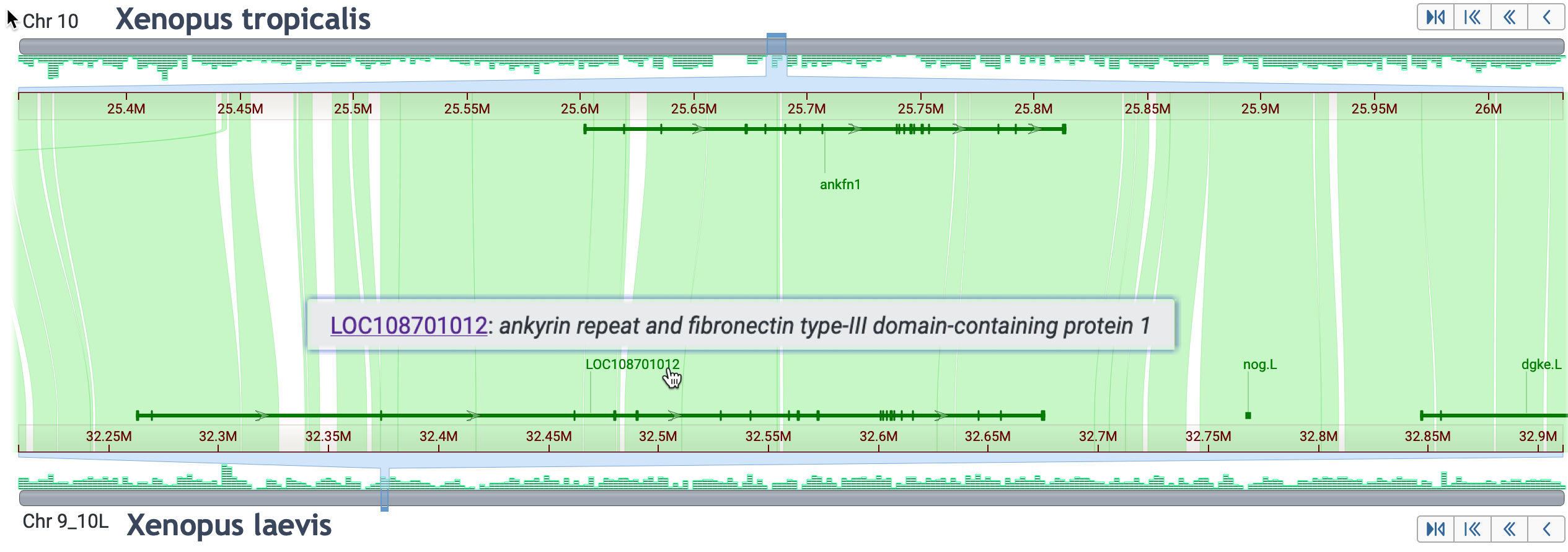
Skills test #1: NCBI Orthologs does not list an ortholog in X. laevis for the gene ankfn1, ankyrin repeat and fibronectin type III domain containing 1; however, there is an ortholog in X. tropicalis. See this Datasets Gene page:
https://www.ncbi.nlm.nih.gov/datasets/gene/id/4a26a75cab8d8cb6d8566dfc1727f140/
Find a candidate for ankfn1 in X. laevis.
Click here to reveal the answer
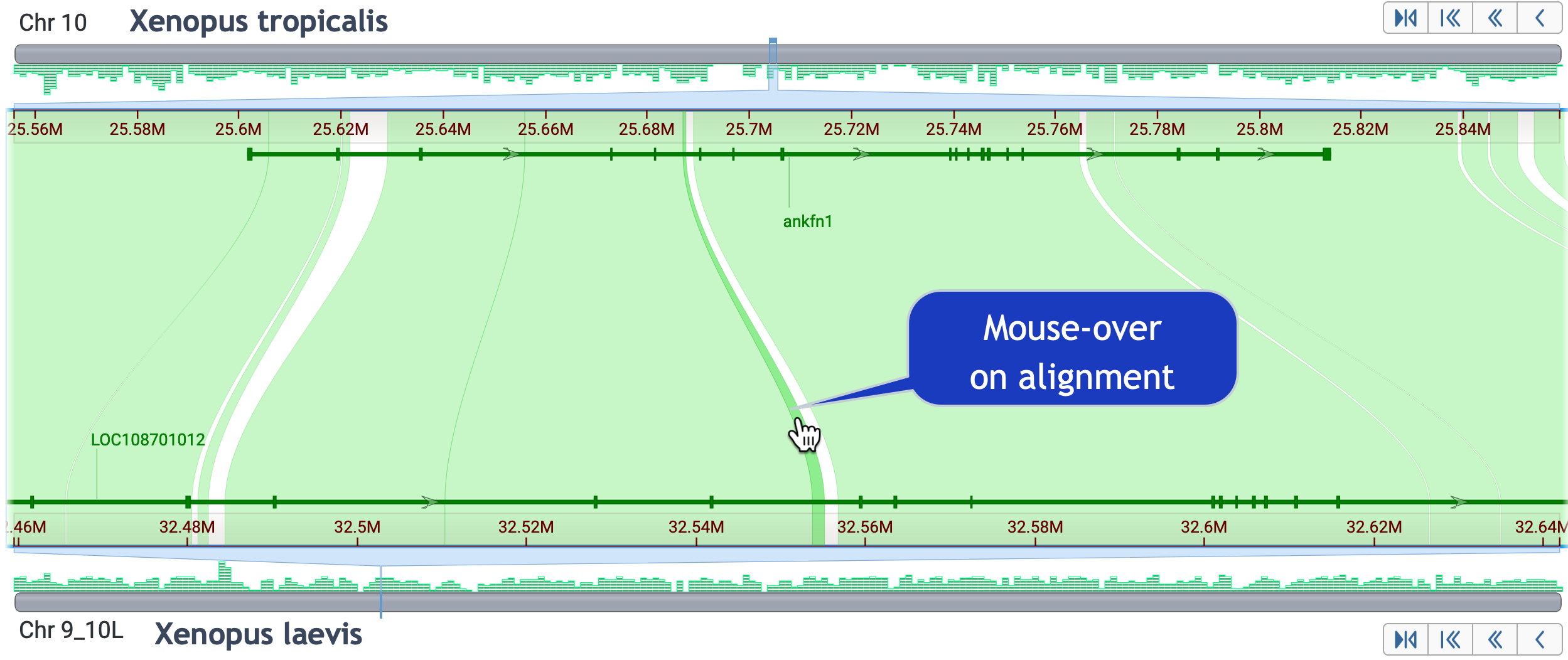
Skills test #2: You can capture nearly all of ankfn1 on X. tropicalis in multiple alignments. How many alignments does that take?
Click here to reveal the answer
Adjust the view to include non-best placed alignments, and set the minimum alignment size to 1k. By mousing over individual alignments, you find that 9 alignments are required to span the complete gene region; two alignments are very small.
Skills test #3: Explore gene order synteny conservation around the ankfn1 like gene. Include the nog2 gene.
Click here to reveal the answer
Here is the default view of the ankfn1 like gene region: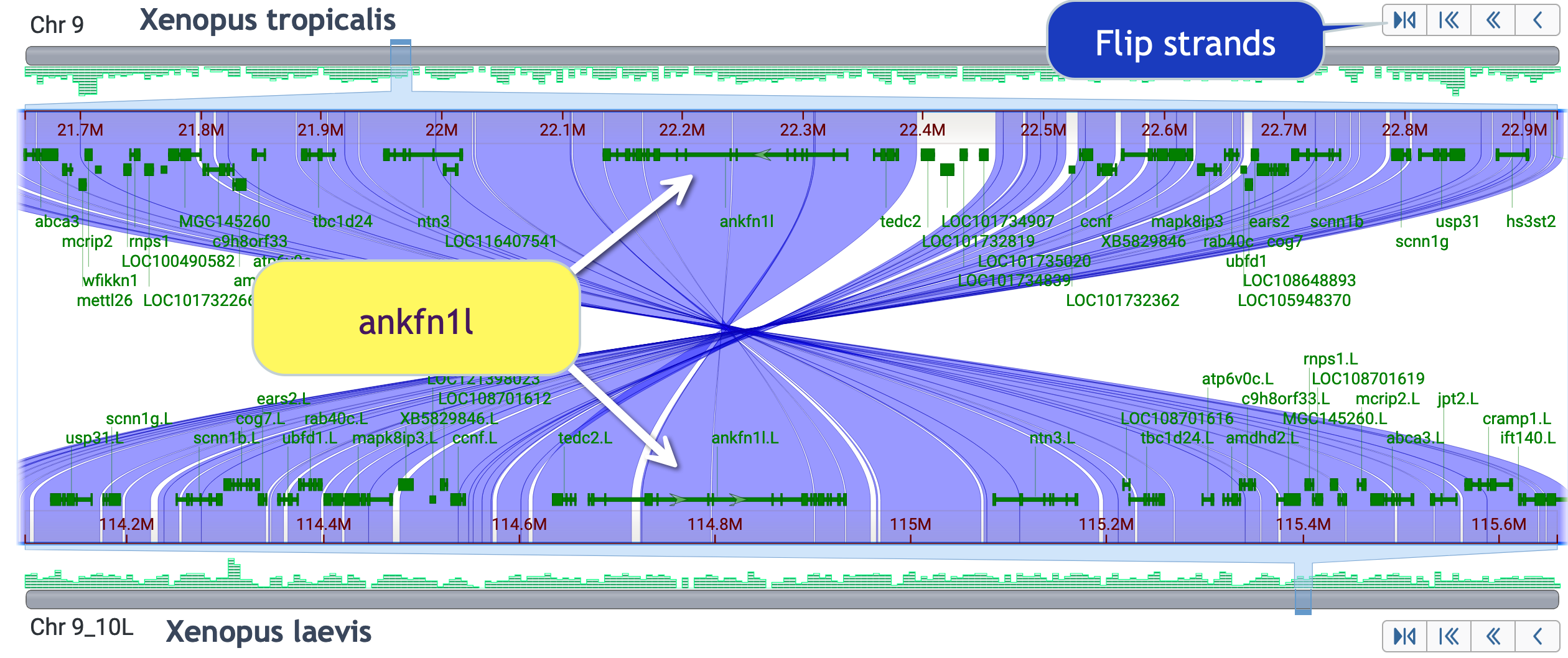
Use "flip strands" to make this easier; and zoom in enough to see the small nog2 gene.
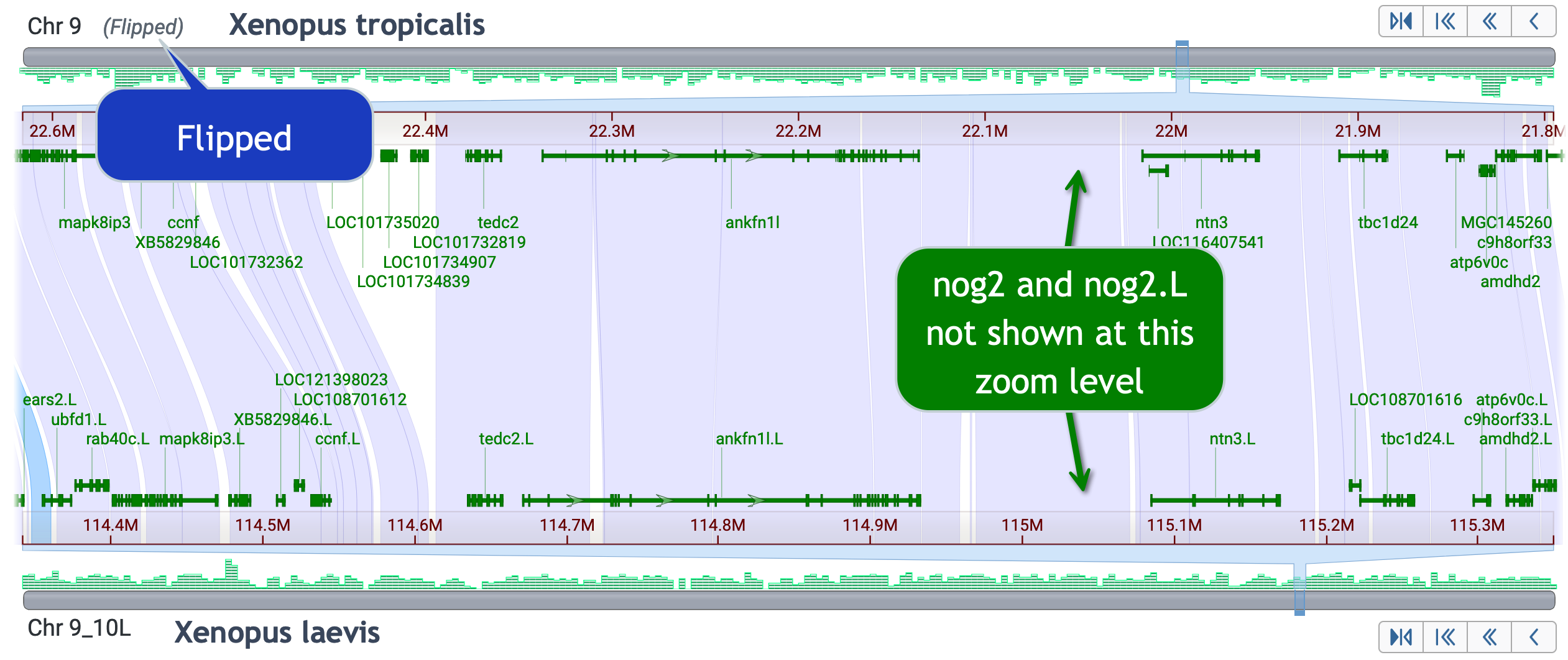
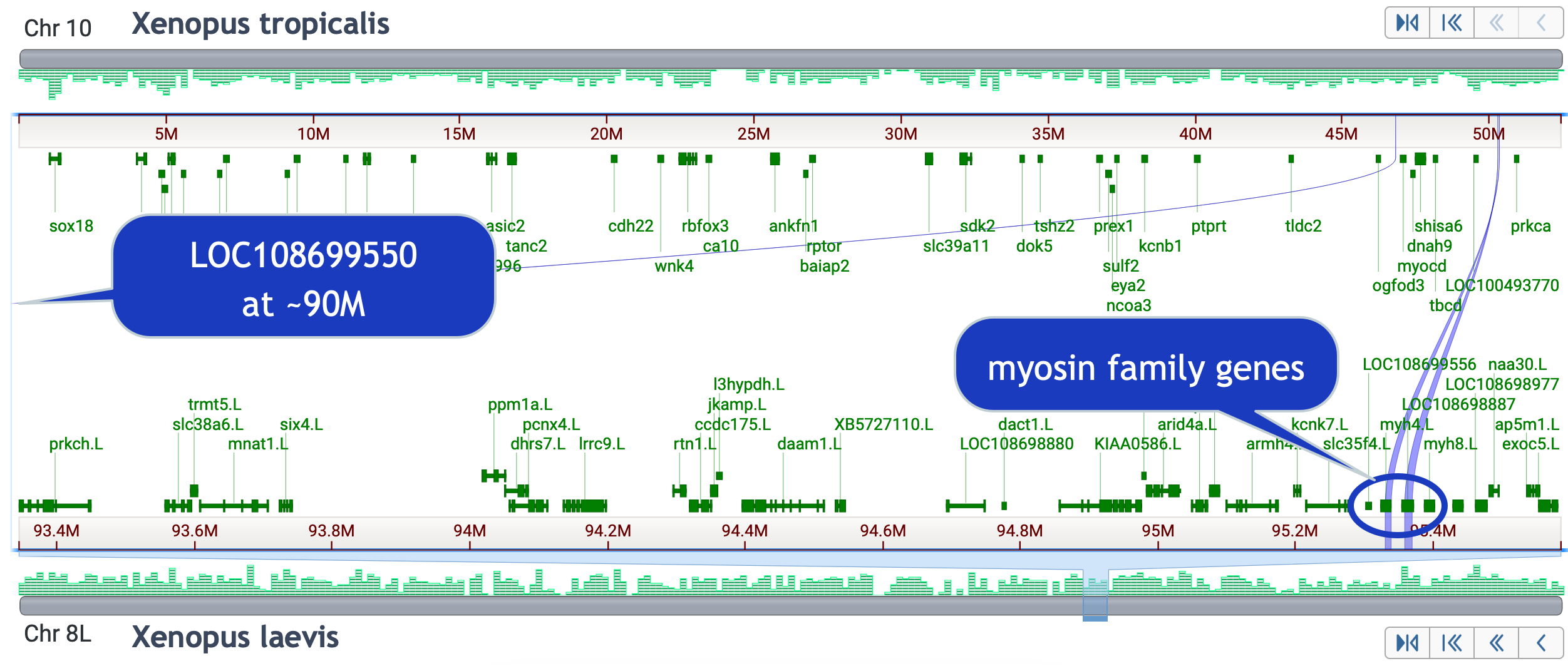
Skills test #4: Find genes annotated on X. tropicalis chr10, that are not on X. laevis 9_10L or 9_10S.
Click here to reveal the answer
- Click on chr 10 in X. tropicalis
- Adjust the view to include non-best placed alignments
- Click on chr 8L and locate the aligned regions
Page 1 of 1




