Case Study: Jeff

Welcome to Your Patient!
Researching the Referral
To learn more about a case, please click on the Referral icon to open the form.
In the Patient Case Review guide sheet provided by your attending: Notate some key clinical features listed for Jeff as well as a preliminary diagnosis.
This is the beginning of preparing for a possible molecular case presentation.
cirrhosis AND diabetes AND ascites AND splenomegaly
 If you need it, you can click here to get to a direct link for the Medgen record page.
If you need it, you can click here to get to a direct link for the Medgen record page.
The "Disease Characteristics" section of this record displays a summary from a specific GeneReviews® Chapter on the NCBI Bookshelf.
Scan the summary and consider accessing and reading the entire GeneReviews® Chapter!
In the Patient Case Review guide sheet: Notate your preliminary diagnosis.
What other disorders might you consider in your differential assessment?
Another good source for information about related disorders is in the “Term Hierarchy” section of this record. Here you can see a few sub-types of Hemochromatosis. Click the names of the disorders to open the MedGen records to read about each sub-type. Note at the top is a gene or genes that have been associated with the disorder.
 You discuss the possible diagnosis with Jeff. He has heard that this is may be a genetic condition. He has a twin sister and a brother and is concerned that they may be affected. He’d like to have a DNA test to see if the cause of his disorder is genetic and also see if he can find out specifically what is wrong with him on a personal level, even though it may not change his prognosis or therapy. You discuss the possible diagnosis with Jeff. He has heard that this is may be a genetic condition. He has a twin sister and a brother and is concerned that they may be affected. He’d like to have a DNA test to see if the cause of his disorder is genetic and also see if he can find out specifically what is wrong with him on a personal level, even though it may not change his prognosis or therapy.You decide to move forward with ordering a genetic test and send an order to the Genetic Testing Laboratory. |
What does the patient's genetic test report say?
Jeff went to the laboratory, provided a sample which was analyzed and the results have been sent to you for consultation. To see Jeff's genetic test results, please click on the Test Results.What is currently known about the identified variant?
Genetic testing laboratories attempt to stay up to date with what is known about the genetic variants that they are assessing. However, it is sometimes valuable to quickly consult with national database of clinical variants (NCBI's ClinVar database) to learn what other organizations have asserted/interpreted for that variant, if anything. In addition to information from testing laboratories, ClinVar receives curated interpretations from authoritative sources such as ClinGen, ACMG and disorder-specific specialist panels.Look over the ClinVar records to see information submitted to NCBI's ClinVar from clinical laboratories, expert panels and clinical genetics organizations.
Look in the Variant Details tab to find biology-related information about this variant.
What is currently known about the identified gene?
If a particular gene has been implicated in a genetic test results indicating a pathogenic variant exists in a patient, it is often helpful to understand what that gene is, what its normal function is, where it is found (cellular and tissue expression patterns), and other sources of accessible information, such as links to relevant scientific literature.
NCBI's Gene database aggregates data from many NCBI databases as well as other high-quality resources to provide information and links to help users find and understand what is currently known about a particular organism's gene.
To learn more about Jeff's impacted gene, search NCBI's Gene database with the gene symbol indicated on the genetic test result and find the record for the human version.
What is the normal function for the gene product? What else has been noted?
In the Patient Case Review guide sheet: Notate what you feel is important to know about the normal version of this gene product.
In which tissues is this gene normally expressed?
In the Patient Case Review guide sheet: Notate the tissues in which you would expect this gene to play a physiological role.
In the Gene Ontology Section of this record is a set of annotations for where this gene product is likely to be found within a cell (Component), what processes it is often involved in (Process), and what it does (Function).
Does this make sense based on the Gene Summary of the Gene that you found above?
Let's put this all of this together....
Does what you've found above make sense based on the patient's symptoms and personal history?
Click here to see some additional helpful information I've pulled it together from an old molecular biology textbook available on the NCBI Bookshelf!
Iron transport and the Transferrin Cycle:
|
Map the variant through the bioinformatic flow!
Now that we understand which gene may be affected by the presence of the detected variant, mapping the variant through the central dogma of molecular biology can help indicate at which point it has its strongest impact.
 Click here to review an overview of the central dogma and genetic variation.
Click here to review an overview of the central dogma and genetic variation.
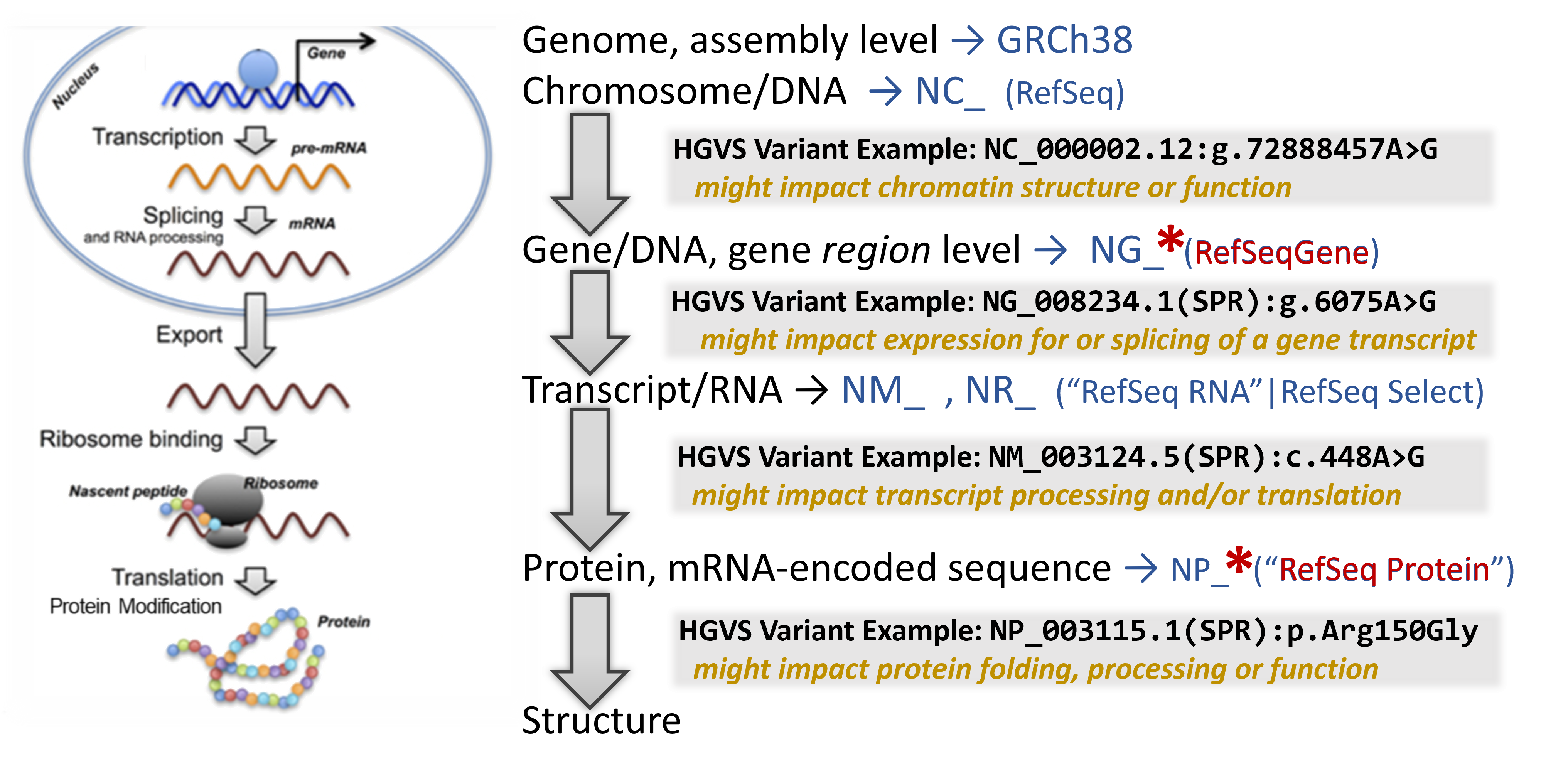
In addition to a lot of helpful aggregated information, NCBI's Gene database provides links to visualization tools which can help to identify where a variant is located in several critical biomolecules.
To learn about the molecular impact of the genetic variant, begin your search on the relevant NCBI Gene record. Then sequentially click on several helpful linked resources on this page to map the location and infer it's impact through this bioinformation flow
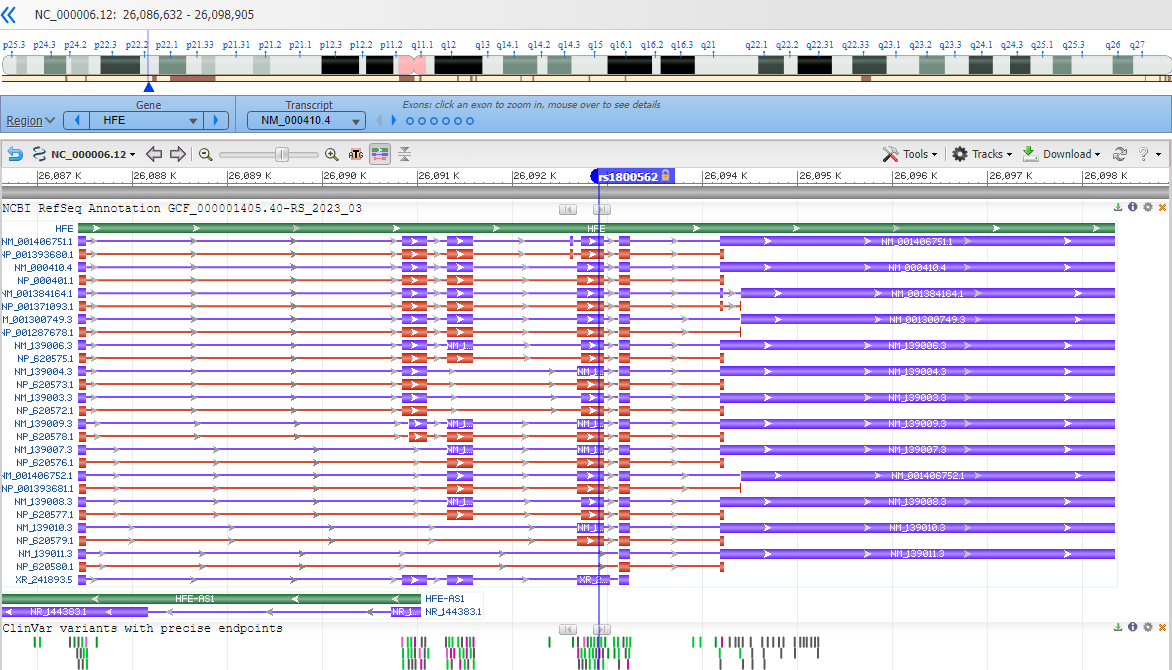
Note that you are looking at a portion of the chromosome (the accession shown is an NC_).
Which chromosome do you think you are looking at? i.e. which one is this gene encoded on?
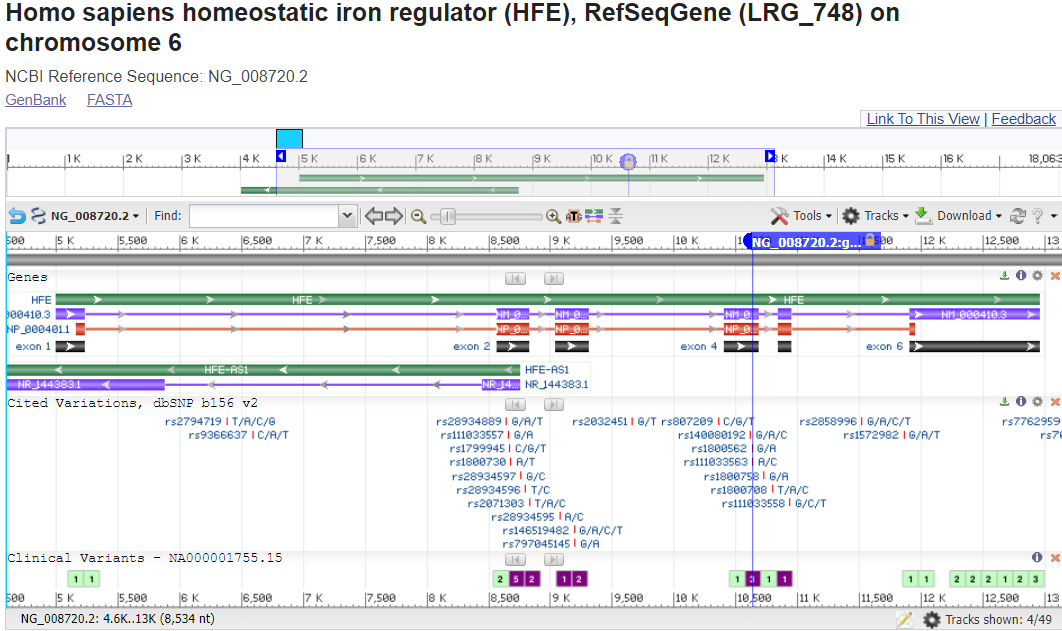
Zoom into the location of the variants by entering an HGVS term, such as: NG_008720.2:g.10633G>A into the top-left text box to search for the variant location.
Where is the variant located in relation to the gene structure? (in the upstream or downstream region surrounding the gene? in the 5'-untranslated portion of the transcript portion? near a splice site? in a coding exon?)
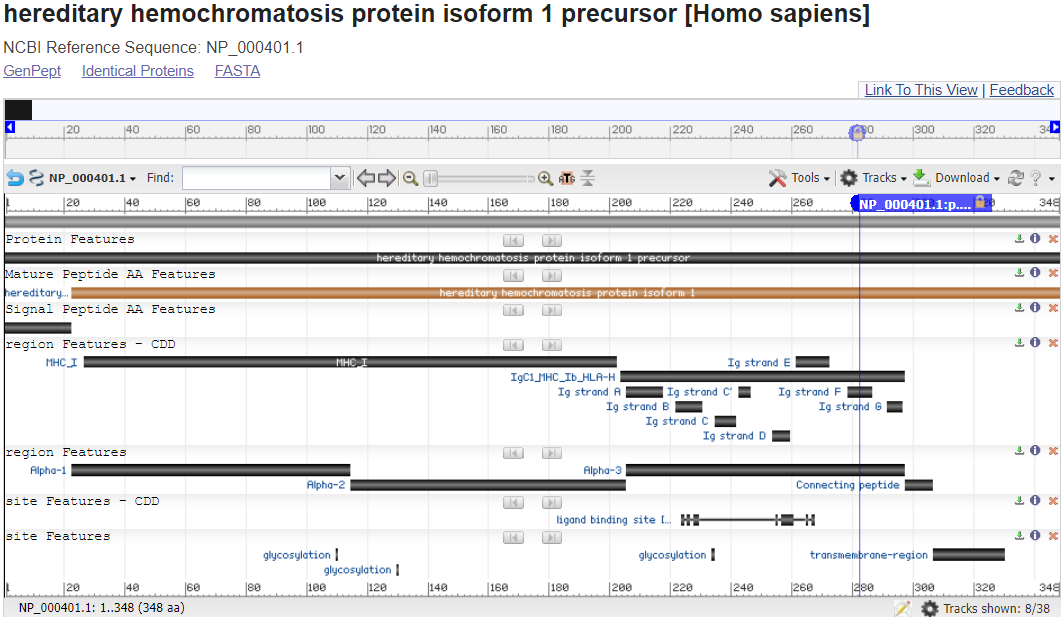
Note that you are looking at the full length of the protein sequence itself in this view. (the accession shown is an NP_).
For many gene products such as this one, you can even see the 3D Structure of the protein, sometimes substrates and ligands and sometimes full complexes and map the location of the genetic variant impact within it.
 Click here to see something pretty cool!
Click here to see something pretty cool!
 The 3D crystal structure for the HFE Protein
The 3D crystal structure for the HFE Protein(PDB accession: 1DE4) in NCBI’s Cn3D Viewer.
The Cys282 position is shown in a yellow and participates
in a disulfide bond (with another cysteine side chain,
orange stick) holding together two beta sheets in an Ig
Fold.
What would the change of that cysteine do to the structure?
What might be the impact on the ability of this protein to function?
To understand the role of the HFE protein in biology, let’s look at two other 3D Structures that arerelated to this protein’s function and purpose (PDB accessions 1SUV – Transferrin Receptor complexed with Ferro-Transferrin & 1DE4 – Transferrin Receptor complexed with the HFE protein).
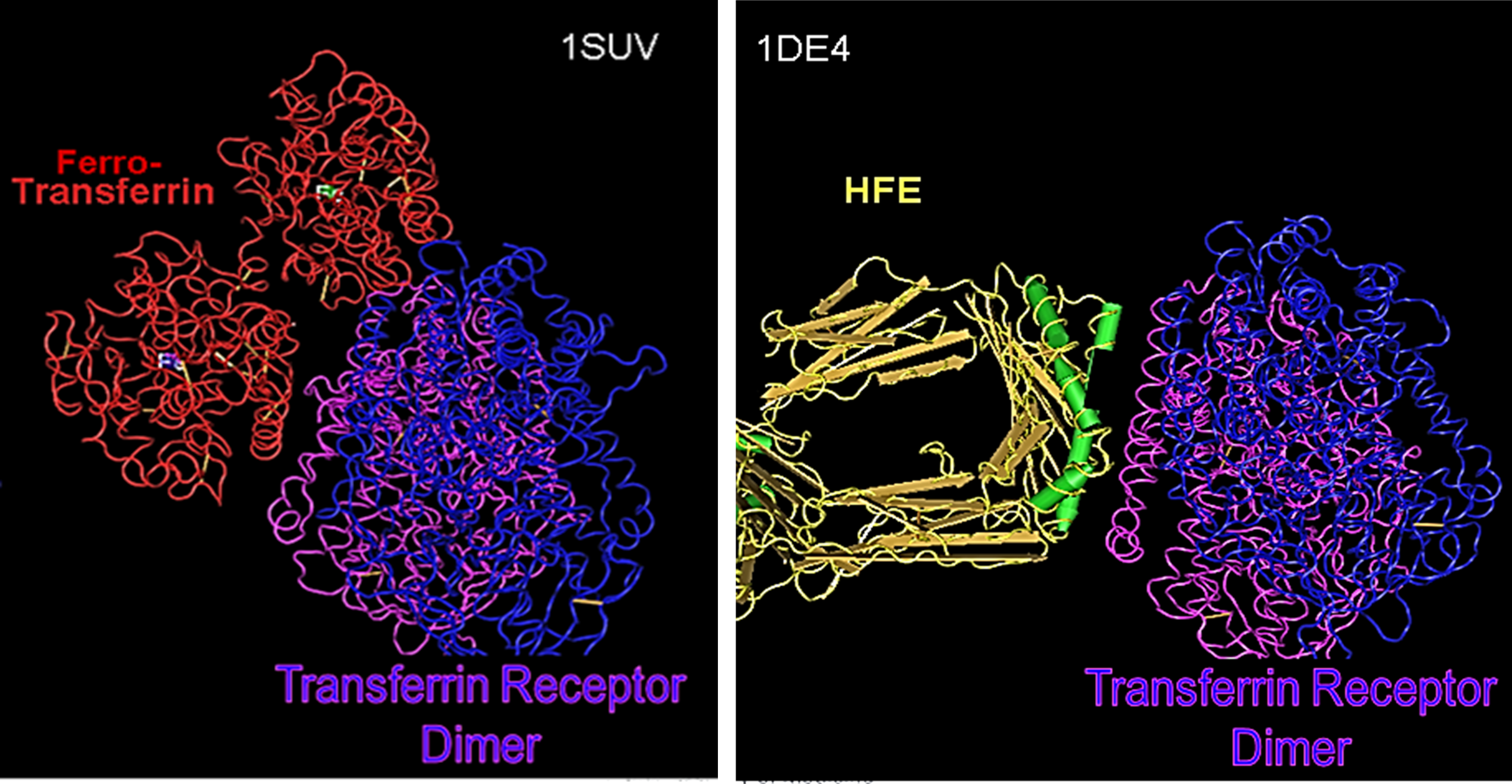
In both structures, the Transferrin Receptor is shown in purple & blue. On the left in red, you see how it complexes with Transferrin (ferro-transferrin means it is carrying its Iron ion payload). On the right in yellow, orange & green (secondary structures – which are also shown in the picture of #9), you see how the Transferrin Receptor complexes with a homodimer of the HFE protein.
How do you think HFE regulates the interaction of the Transferrin Receptor with Ferro-Transferrin?
What would happen to this if the HFE protein is encoded with the p.Cys282Tyr variant?
Let's put it all together to understand what is happening in the patient!
 Jeff would like to have some answers.
Jeff would like to have some answers.
Click here to review some things you may want to consider when formulating the answer to his questions.
-
-
-
-
- Which gene is impacted by the genetic variation and what does the gene product normally "do"?
- what is it's biomolecular function?
- what is it's impact on cellular physiology?
- in which cells/tissues is the gene product usually expressed?
- Based on the patient's variation(s):
- what do you think this would do to the gene product's structure and biomolecular function?
- what would this do to cellular physiology?
- what tissues or organs impact be impacted?
- Based on the proposed impacted-tissues/organs, may some of the the patient's symptoms be explained by this? (validating her experience)
- Which gene is impacted by the genetic variation and what does the gene product normally "do"?
-
-
-

What do you think is happening in your patient?
| Notes | |
| Diagnosis | |
| Genetic Variation(s) | |
| Proposed Molecular Mechanism of Variant Impact | |
| How does this relate to the phenotype? |
Take-away message!
Workflow: We've practiced this same step-by-step process to learn more about a different patient's genetic variant.
Genetic disorder & molecular pathology: Missense variants involving cysteine residues, in particular, can have a devastating effect due to their key role in forming structure-stabilizing, covalent disulfide bonds across spans of a protein sequence. The loss of this structural cross-beam support can cause the protein to unfold, often disrupting at least part of the protein's ability to function and sometimes triggering an endogenous "unfolded protein response" which activates a process to destroy the unnaturally unfolded protein.
In this case, the HFE protein is destroyed and with it, it's ability to bind to and inhibit the Transferrin Receptor's import of Ferro-transferrin. Thus, the cellular import continues unregulated and the cell accumulates dramatically increasing levels of highly reactive iron ions. Over time, these cause damage to the cell and if it is not reversed and/or if the cell cannot undergo apoptosis (a clean, programmed cell death), then significant DNA and protein damage can accumulate and induce the cell to undergo neoplastic transformation. The most notorious cancer eventually developed by those with Hemochromatosis, being hepatocellular carcinoma.
ANSWER
Last Reviewed: August 22, 2023





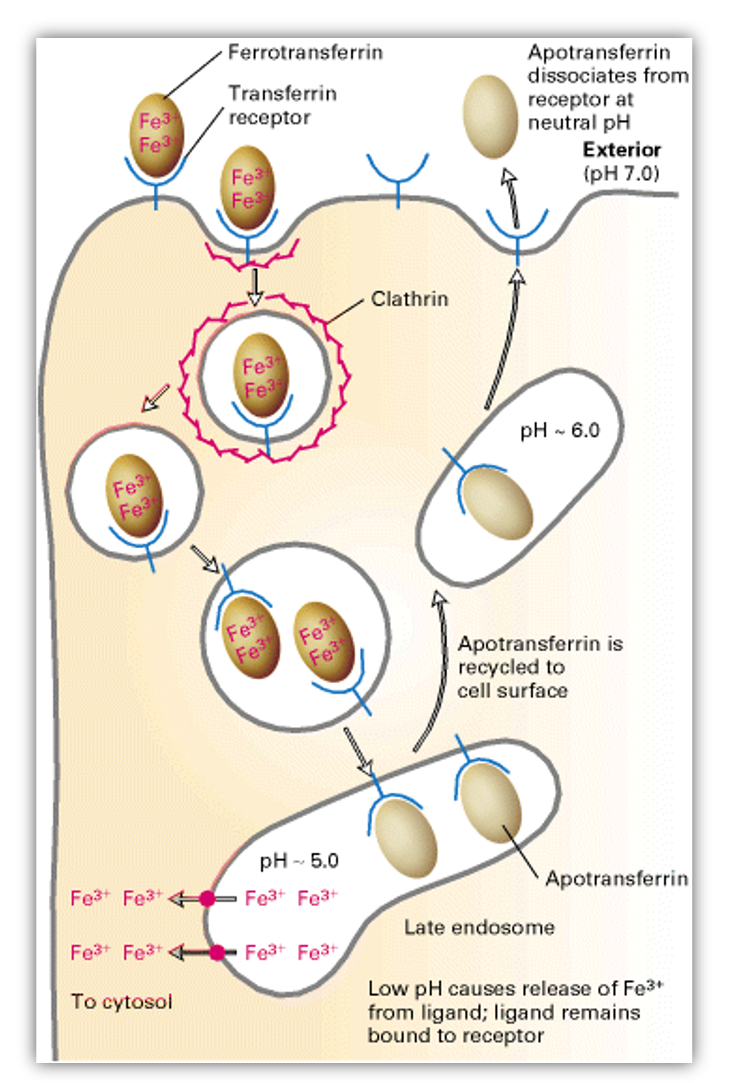 Iron is an essential nutrient required for the synthesis of hemoglobin, cytochromes and many other proteins.
Iron is an essential nutrient required for the synthesis of hemoglobin, cytochromes and many other proteins.