Step 5: Searching for sequence orthologs using BLAST
Task: identify orthologs of IMA1 in S. pastorianus and S. eubayanus.
Now we have a high-quality RefSeq protein sequence for the IMA1 gene in S. cerevisiae! The genomes of the other two yeast species have less information, like gene names, associated with their genome assemblies. So, instead of searching for IMA1 by name, we search for proteins that have similar actual sequences to the S. cerevisiae protein sequence in the available NCBI sequence data for S. pastorianus and S. eubayanus.
We will use an NCBI program called BLAST (Basic Local Search Alignment Tool) to search for orthologs of the IMA1 in S. pastorianus and S. eubayanus.
Background on NCBI Resources Used:
NCBI BLAST - The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to retrieve similar sequences with informative metadata to infer the source organism for the isolate, identify potentially related members of gene families, as well as explore evolutionary or functional relationships between sequences. In this case, we are using BLAST to look for sequences similar to the IMA1 protein in specific databases.
To run a BLAST search, we need to:
- Choose a specific BLAST program
- Supply a query sequence
- Choose the set of data we want to search
- Adjust any other parameters (advanced)
- Run the search!
Once we have the results, we can:
- Select and filter results to examine further
- Save or download relevant parts of the results output
- View multiple sequence alignments of the results to identify interesting changes
- Summarize the output in many other ways!
You will learn how to complete these tasks as relevant for our goal.
Setting Up the BLAST Search
1. Choose and load a query sequence: For us, this will be quick. To get started, go directly to a BLAST search with the S. cerevisiae protein sequence already loaded by clicking “"Run Blast" Under the "Analyze" menu on the RefSeq protein page.
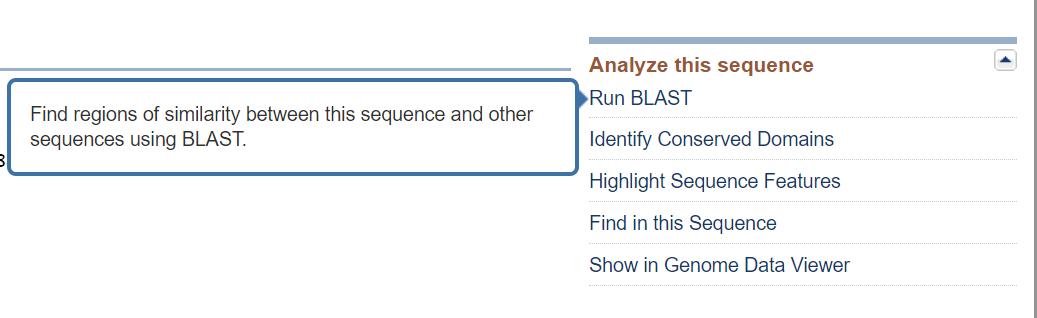
This will take you to a BLAST interface page with the accession number for the RefSeq protein already listed in the Entry Query Sequence Box. 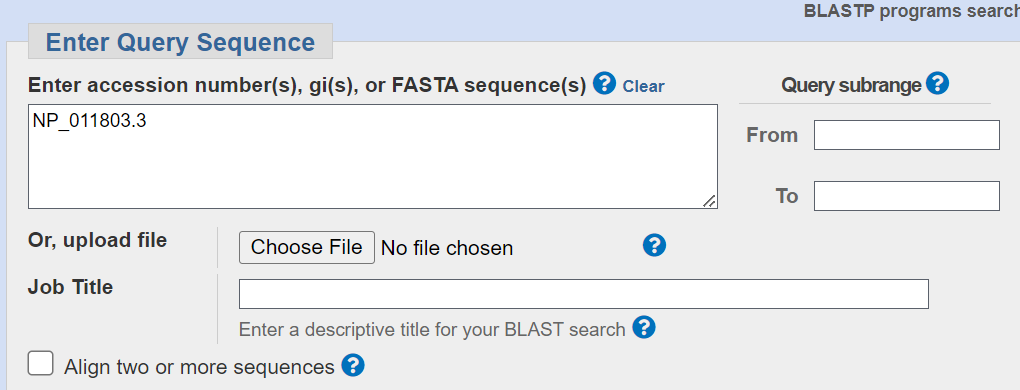
You could also have copy and pasted another accession number, the actual DNA sequence or uploaded a file of accessions or actual sequences. Many more details about acceptable input formats can be found on this BLAST Topics: Query Input page.
2. Choose the appropriate algorithm for your BLAST search: This is already taken care of for you by following the link from RefSeq! Looking above the query input box, you will see that “blastp” is selected.
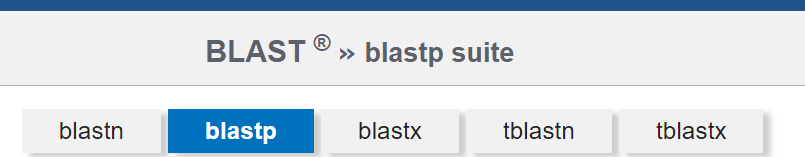
The BLASTP algorithm is designed to search through protein databases using a protein sequence query, perfect for your purposes here, since we will be searching against the NCBI Non-Redundant Protein Database (NR). You don’t need to modify anything here.
Depending on the type of input query and database you have available, and the types of research questions you have, you may choose a different BLAST program:
- Nucleotide BLAST (blastn) searches a nucleotide against a nucleotide database
- Protein BLAST (blastp) to search a protein against a protein database
- blastx translates a nucleotide into a protein and searches it against the protein databases
- tblastn searches a protein query against a translated version of the nucleotide databases
- tblastx searches a translated nucleotide query against a translated version of the nucleotide database.
For example, if we wanted to search a whole transcriptome (mRNA) dataset with this protein query, we would probably want to use tblastn. You can choose one of these programs directly from the web BLAST home page.
3. Choose a database to search: In this step, we are going to specify that we want to search a subset of the NR database containing S. pastorianus and S. eubayanus data.
To start:
- Leave `Standard Databases` selected
- Leave Database set to “non-redundant protein-sequences”
Hovering over the question mark symbol, we can see that as of June 26th, 2023, there are over 572 million sequences in NR!
Narrow that search down to just our species of interest, using the Organism menu:
- In the box, start typing `Saccharomyces pastorianus`
- Select “Saccharomyces pastorianus (taxid:27292)”
- Click “Add Organism”
- Search for Saccharomyces eubaynus
- Select “Saccharomyces eubayanus (taxid:1080349)”
Your options should look like this when you are done:

Identifying Relevant Info from BLAST results
Click here if you need to jump to the BLAST results page: Saved BLAST Results for S. cerevisiae IMA1 protein
1. Getting information from the search summaryAmong other useful information, the top section of a BLAST results page can help you check that the BLAST search went as planned:
- Program is blastp
- Database was NR
- Our query ID matches our input accession number
- IMA1 is in the description
- Our search was was limited to only the two yeast species
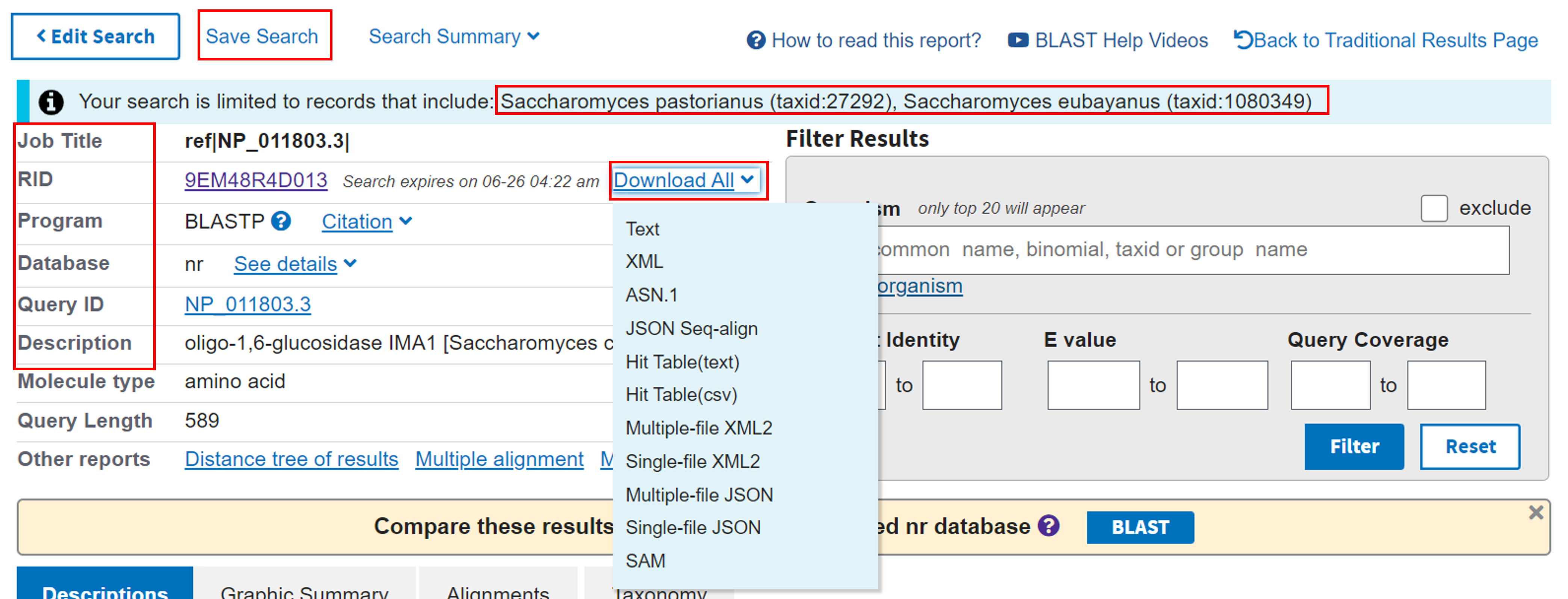
Temporary result-sharing option: The BLAST results are only stored by NCBI for 36 hours. If you would like to send your own results page to someone within that time period, copy the link you get from clicking on the RID. We have specially saved a permanent version of the search result for this example at this link here.
Optional advanced option: As BLAST results are only stored on the NCBI server for a limited time, click “Download All” (see menu in above figure) to save the actual results of this exact BLAST search in a number of formats that can be used for further analyses.
2. Save your search strategyBecause the actual results of the BLAST search are only preserved on NCBI for a short period, you can save the search strategy to run this exact search again.
- Click “Save Search”
- This will direct you to a page “Saved Search Strategy page
- Click on the “View” button in an entry under “Saved Search Strategies”
- This will load a BLAST interface with all of the same options as for our search above so you can run it again.

3. Examine results to find orthologs from the other yeast species
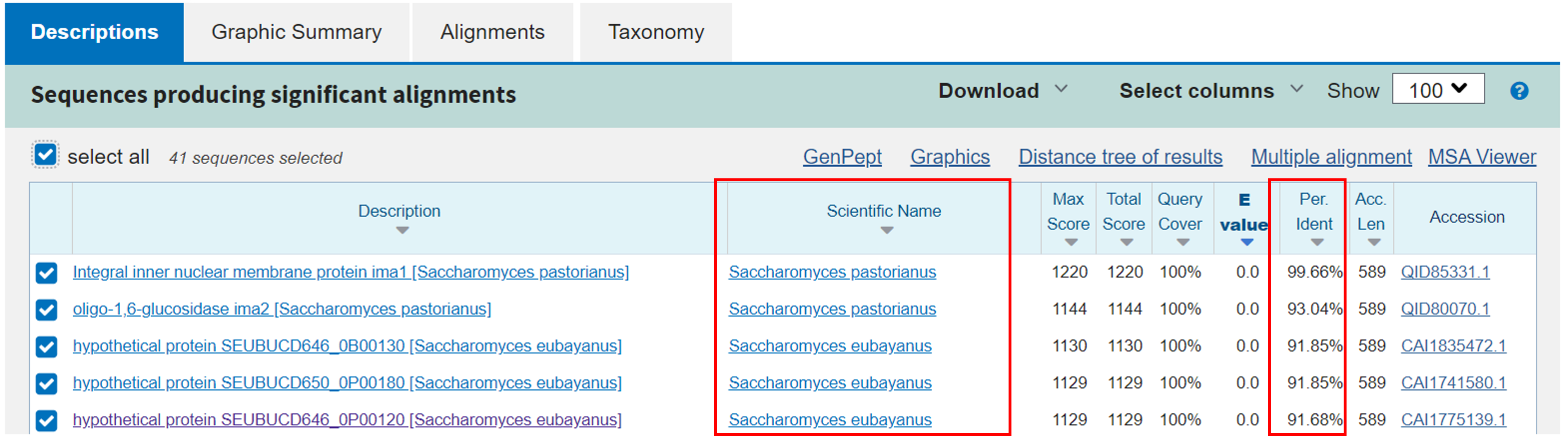
Scroll down to look at the actual IMA1-like sequences that BLAST identified in the S. pastorianus and S. cerevisiae data. 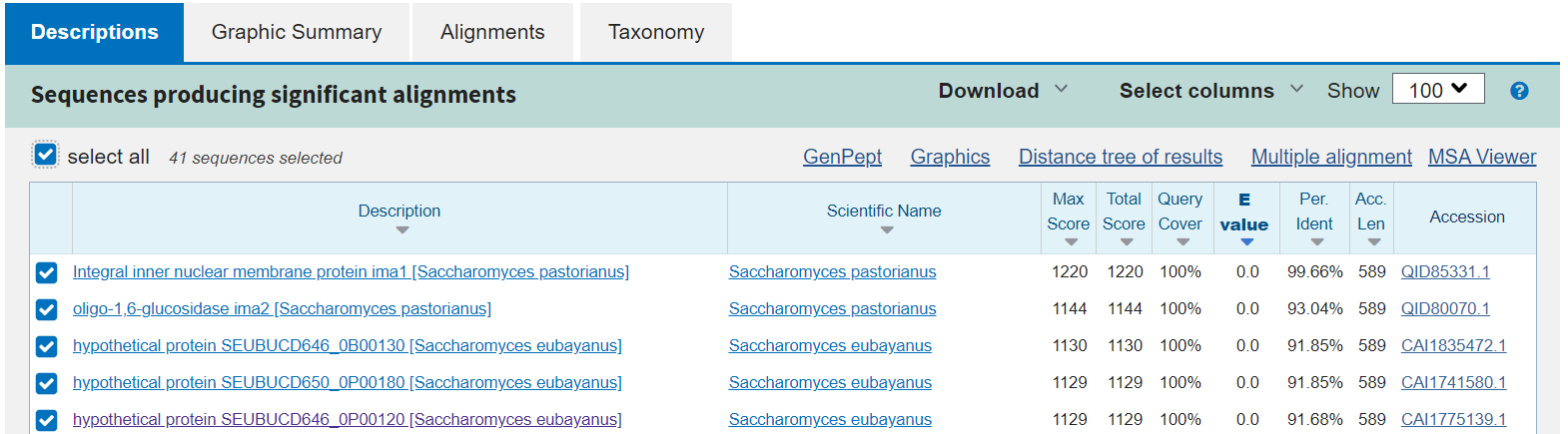
Each row under “sequences producing significant alignments” is a protein sequence significantly similar to the S. cerevisiae IMA1 protein query. At the time of the workshop, there were 41 of these results. Discussing each of the columns in this table is outside the scope of this course, but there are a few things to note.
The following columns link to further details about each result:
- Description: Clicking name will take you to an alignment of this protein to the query.
- Scientific name: Link takes you to the relevant Taxonomy page.
- Accession number: Links to NCBI Protein Database page for subject
The rest of the columns help you decide which results best match the input query sequence. A few of them you might want to look at are:
- Query coverage: Percent of the query that aligns with the database entry/subject sequence
- Per. Ident. (percent identity): Percentage of base pairs that are the same between your query and the result
- E-value: A statistical measure of whether this match could have occurred by chance, lower numbers mean more significant! Read a bit more about E-values in the BLAST FAQ page.
What do you see? Are there potential orthologs of IMA1 in S. pastorianus and S. eubayanus?
 For a hint about what to look for, click here.
For a hint about what to look for, click here.
Step 5 Conclusions
| Main takeaway: We can use BLAST to find orthologs of a gene/protein using a sequence-similarity approach, even in species without well-annotated genomes! |
In this exercise you learned how to:
|
Further BLAST Resources:
- BLAST Help Youtube Page
- BLAST results page walkthrough flyer
- Upcoming "Exploring Evolutionary Relationships Using BLAST" workshop - applications due July 25th!
Last Reviewed: June 28, 2023

